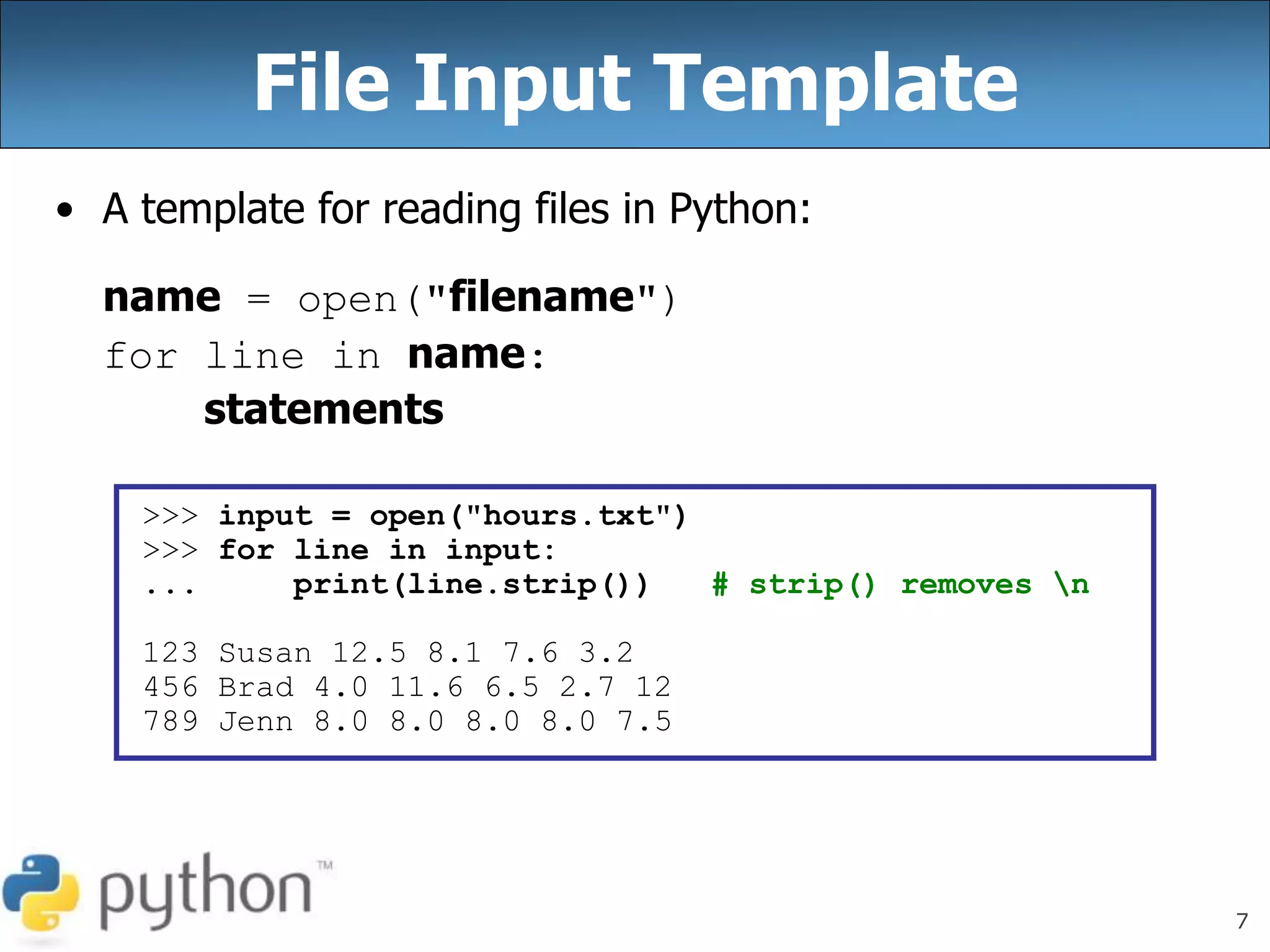
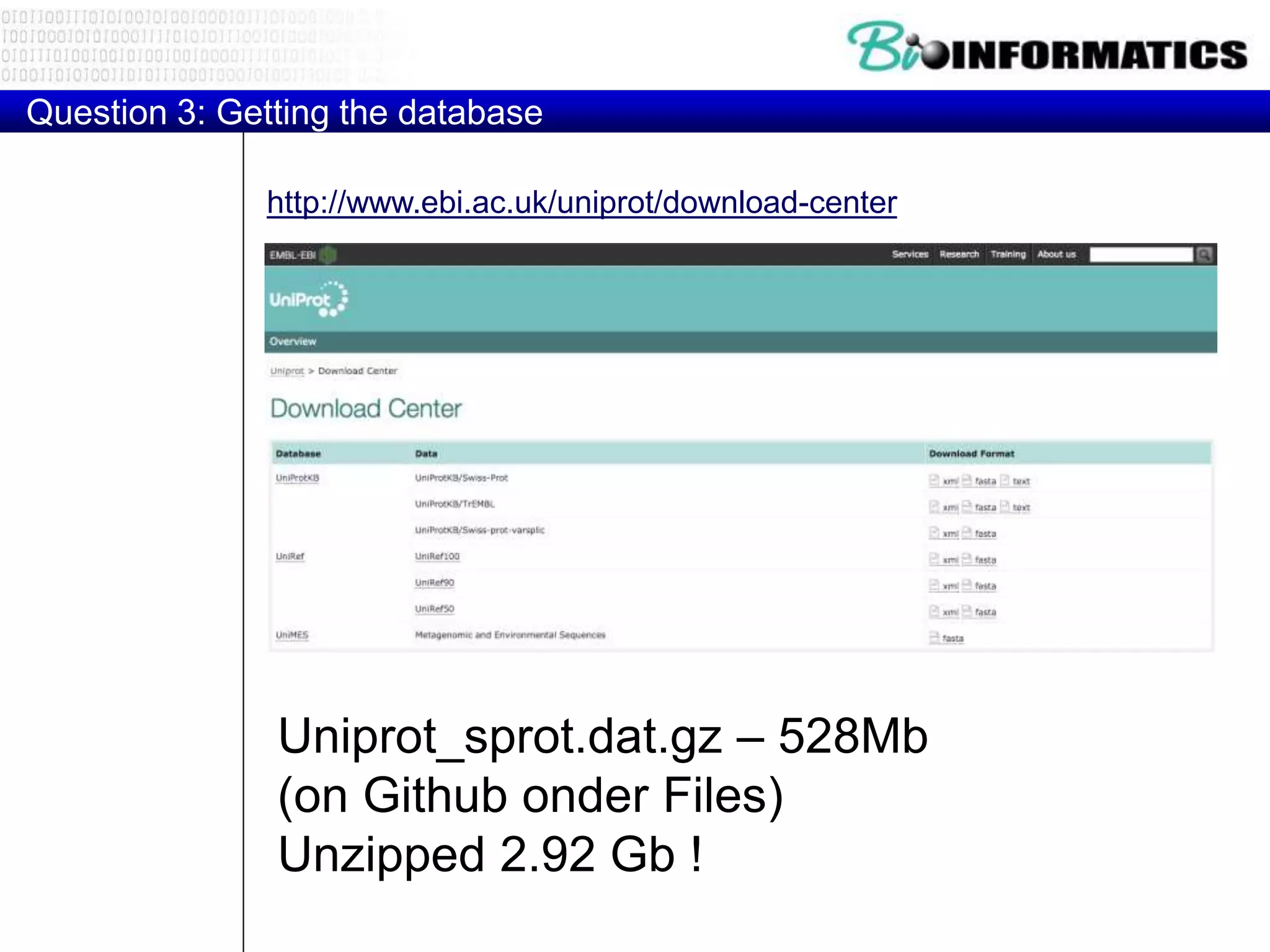
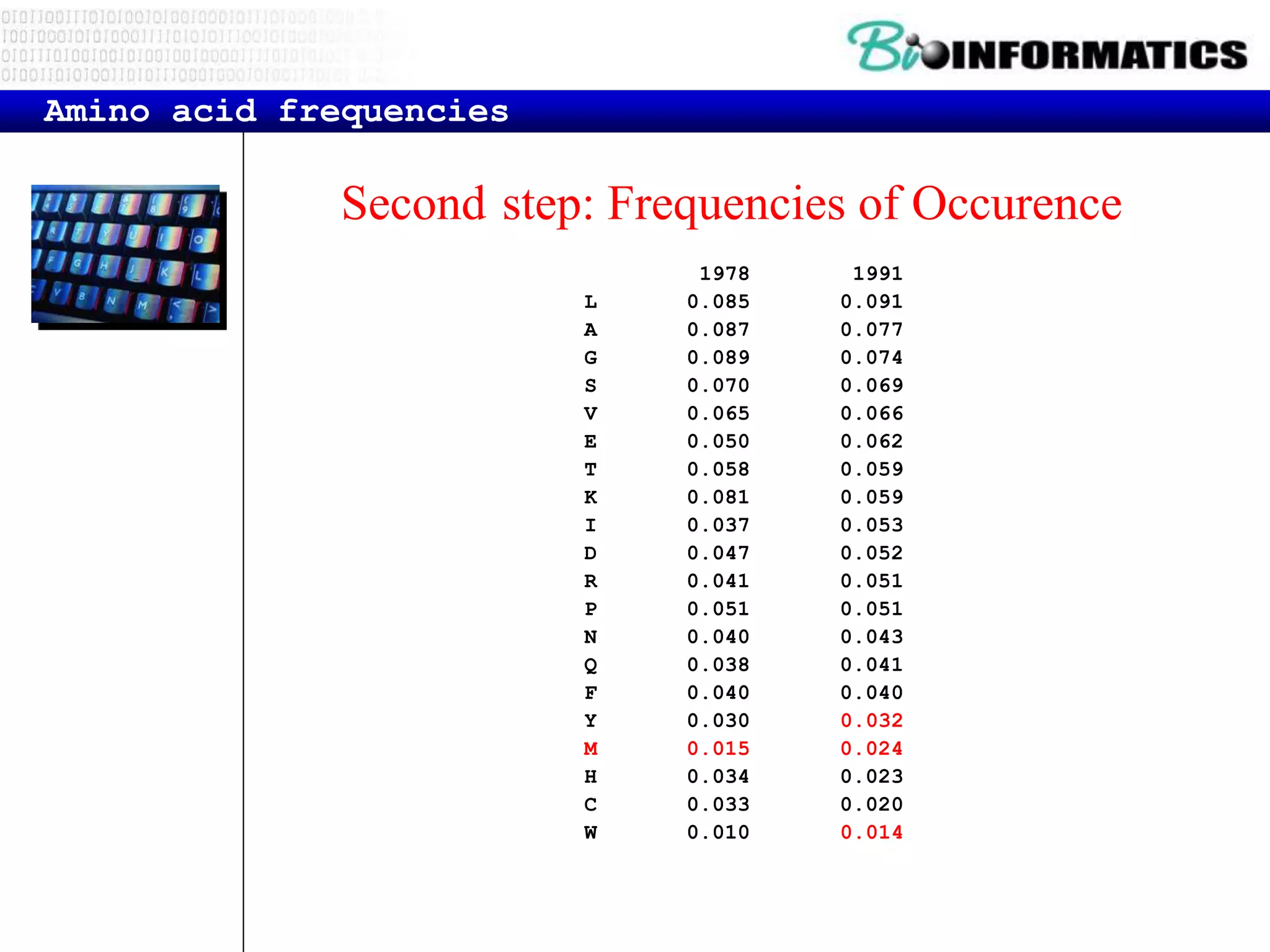
The document discusses reading and writing files in Python. It provides examples of opening files for reading, writing, and appending. It demonstrates how to read an entire file, individual lines, and loop through lines. It also shows how to write strings to files and close files once writing is complete. Additional topics covered include a template for reading files line by line and examples of counting lines, words, and characters in a file.


![Recap
if condition:
statements
[elif condition:
statements] ...
else:
statements
while condition:
statements
for var in sequence:
statements
break
continue
Strings
REGULAR EXPRESSIONS](https://image.slidesharecdn.com/2016bioinformaticsiiowimvancriekinge-161107233350/75/2016-bioinformatics-i_io_wim_vancriekinge-4-2048.jpg)