This document provides an overview of using Python for bioinformatics. It discusses why Python is useful for bioinformatics due to its built-in libraries and wide scientific use. It also covers Python basics like strings, regular expressions, control structures, lists, dictionaries, reading/writing files, and using GitHub for code sharing. Examples are given for many of these topics. Finally, it poses questions about analyzing sequence data and a protein database using Python.








![Run Install.py (is BioPython installed ?)
import pip
import sys
import platform
import webbrowser
print ("Python " + platform.python_version()+ " installed
packages:")
installed_packages = pip.get_installed_distributions()
installed_packages_list = sorted(["%s==%s" % (i.key, i.version)
for i in installed_packages])
print(*installed_packages_list,sep="n")](https://image.slidesharecdn.com/2015bioinformaticspythoniowimvancriekinge-151019212304-lva1-app6891/85/2015-bioinformatics-python_io_wim_vancriekinge-9-320.jpg)
![Control Structures
if condition:
statements
[elif condition:
statements] ...
else:
statements
while condition:
statements
for var in sequence:
statements
break
continue](https://image.slidesharecdn.com/2015bioinformaticspythoniowimvancriekinge-151019212304-lva1-app6891/85/2015-bioinformatics-python_io_wim_vancriekinge-10-320.jpg)
![Lists
• Flexible arrays, not Lisp-like linked
lists
• a = [99, "bottles of beer", ["on", "the",
"wall"]]
• Same operators as for strings
• a+b, a*3, a[0], a[-1], a[1:], len(a)
• Item and slice assignment
• a[0] = 98
• a[1:2] = ["bottles", "of", "beer"]
-> [98, "bottles", "of", "beer", ["on", "the", "wall"]]
• del a[-1] # -> [98, "bottles", "of", "beer"]](https://image.slidesharecdn.com/2015bioinformaticspythoniowimvancriekinge-151019212304-lva1-app6891/85/2015-bioinformatics-python_io_wim_vancriekinge-11-320.jpg)
![Dictionaries
• Hash tables, "associative arrays"
• d = {"duck": "eend", "water": "water"}
• Lookup:
• d["duck"] -> "eend"
• d["back"] # raises KeyError exception
• Delete, insert, overwrite:
• del d["water"] # {"duck": "eend", "back": "rug"}
• d["back"] = "rug" # {"duck": "eend", "back":
"rug"}
• d["duck"] = "duik" # {"duck": "duik", "back":
"rug"}](https://image.slidesharecdn.com/2015bioinformaticspythoniowimvancriekinge-151019212304-lva1-app6891/85/2015-bioinformatics-python_io_wim_vancriekinge-12-320.jpg)
![Regex.py
text = 'abbaaabbbbaaaaa'
pattern = 'ab'
for match in re.finditer(pattern, text):
s = match.start()
e = match.end()
print ('Found "%s" at %d:%d' % (text[s:e], s, e))](https://image.slidesharecdn.com/2015bioinformaticspythoniowimvancriekinge-151019212304-lva1-app6891/85/2015-bioinformatics-python_io_wim_vancriekinge-13-320.jpg)
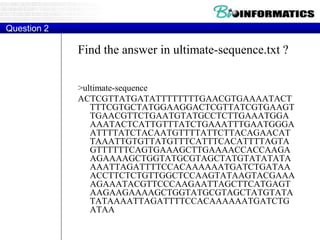

![Hint 2: Translations
Python way:
tab = str.maketrans("ACGU","UGCA")
sequence = sequence.translate(tab)[::-1]](https://image.slidesharecdn.com/2015bioinformaticspythoniowimvancriekinge-151019212304-lva1-app6891/85/2015-bioinformatics-python_io_wim_vancriekinge-16-320.jpg)







