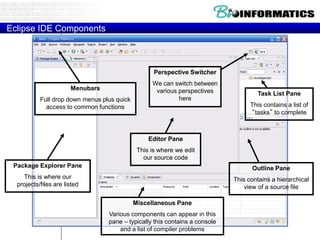
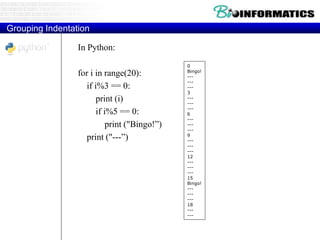
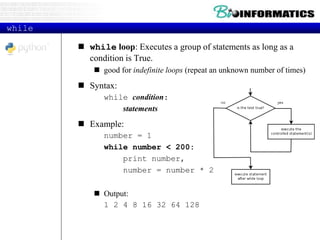
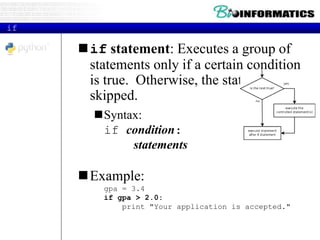
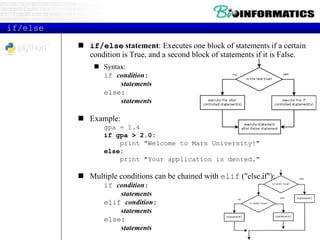
This document provides an overview of Python for bioinformatics. It discusses what Python is, why it is useful for bioinformatics, and how to get started with Python. It also covers Python IDEs like Eclipse and PyDev, code sharing with Git and GitHub, strings, regular expressions, and other Python concepts.





![Run Install.py (is BioPython installed ?)
import pip
import sys
import platform
import webbrowser
print ("Python " + platform.python_version()+ " installed
packages:")
installed_packages = pip.get_installed_distributions()
installed_packages_list = sorted(["%s==%s" % (i.key, i.version)
for i in installed_packages])
print(*installed_packages_list,sep="n")](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-12-320.jpg)
![Control Structures
if condition:
statements
[elif condition:
statements] ...
else:
statements
while condition:
statements
for var in sequence:
statements
break
continue](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-13-320.jpg)



![Indexes
Characters in a string are numbered with indexes starting at 0:
Example:
name = "P. Diddy"
Accessing an individual character of a string:
variableName [ index ]
Example:
print name, "starts with", name[0]
Output:
P. Diddy starts with P
index 0 1 2 3 4 5 6 7
character P . D i d d y](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-23-320.jpg)
![Strings
• "hello"+"world" "helloworld" # concatenation
• "hello"*3 "hellohellohello" # repetition
• "hello"[0] "h" # indexing
• "hello"[-1] "o" # (from end)
• "hello"[1:4] "ell" # slicing
• len("hello") 5 # size
• "hello" < "jello" 1 # comparison
• "e" in "hello" 1 # search
• "escapes: n etc, 033 etc, if etc"
• 'single quotes' """triple quotes""" r"raw strings"](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-24-320.jpg)

![Lists
• Flexible arrays, not Lisp-like linked
lists
• a = [99, "bottles of beer", ["on", "the",
"wall"]]
• Same operators as for strings
• a+b, a*3, a[0], a[-1], a[1:], len(a)
• Item and slice assignment
• a[0] = 98
• a[1:2] = ["bottles", "of", "beer"]
-> [98, "bottles", "of", "beer", ["on", "the", "wall"]]
• del a[-1] # -> [98, "bottles", "of", "beer"]](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-28-320.jpg)
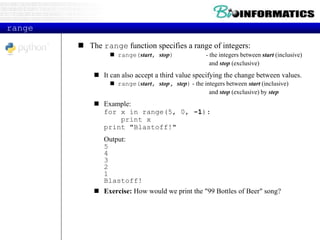
![More List Operations
>>> a = range(5) # [0,1,2,3,4]
>>> a.append(5) # [0,1,2,3,4,5]
>>> a.pop() # [0,1,2,3,4]
>>> a.insert(0, 42) # [42,0,1,2,3,4]
>>> a.pop(0) # [0,1,2,3,4]
>>> a.reverse() # [4,3,2,1,0]
>>> a.sort() # [0,1,2,3,4]](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-29-320.jpg)
![Dictionaries
• Hash tables, "associative arrays"
• d = {"duck": "eend", "water": "water"}
• Lookup:
• d["duck"] -> "eend"
• d["back"] # raises KeyError exception
• Delete, insert, overwrite:
• del d["water"] # {"duck": "eend", "back": "rug"}
• d["back"] = "rug" # {"duck": "eend", "back":
"rug"}
• d["duck"] = "duik" # {"duck": "duik", "back":
"rug"}](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-30-320.jpg)
![More Dictionary Ops
• Keys, values, items:
• d.keys() -> ["duck", "back"]
• d.values() -> ["duik", "rug"]
• d.items() -> [("duck","duik"),
("back","rug")]
• Presence check:
• d.has_key("duck") -> 1; d.has_key("spam") -
> 0
• Values of any type; keys almost any
• {"name":"Guido", "age":43,
("hello","world"):1,
42:"yes", "flag": ["red","white","blue"]}](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-31-320.jpg)

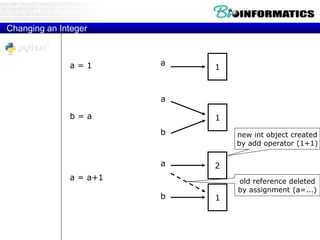
![Reference Semantics
• Assignment manipulates references
• x = y does not make a copy of y
• x = y makes x reference the object y
references
• Very useful; but beware!
• Example:
>>> a = [1, 2, 3]
>>> b = a
>>> a.append(4)
>>> print b
[1, 2, 3, 4]](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-33-320.jpg)
![a
1 2 3
b
a
1 2 3
b
4
a = [1, 2, 3]
a.append(4)
b = a
a
1 2 3
Changing a Shared List](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-34-320.jpg)





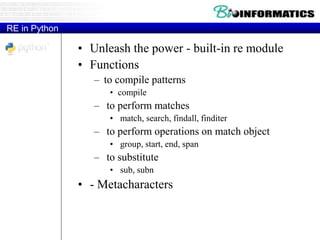
![Quantifiers
• [ATGC]
• You can specify the number of times
you want to see an atom. Examples
• d* : Zero or more times
• d+ : One or more times
• d{3} : Exactly three times
• d{4,7} : At least four, and not more
than seven
• d{3,} : Three or more times
• We could rewrite /ddd-dddd/ as:
– /d{3}-d{4}/](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-42-320.jpg)



![Regex.py
text = 'abbaaabbbbaaaaa'
pattern = 'ab'
for match in re.finditer(pattern, text):
s = match.start()
e = match.end()
print ('Found "%s" at %d:%d' % (text[s:e], s, e))](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-46-320.jpg)



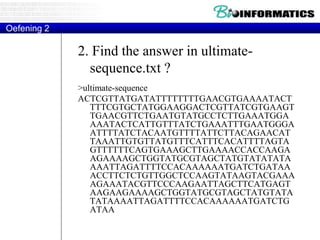
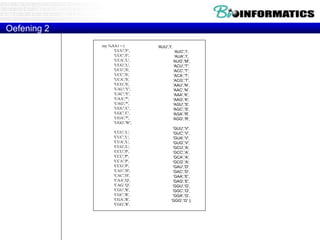

![Translations
Python way:
tab = str.maketrans("ACGU","UGCA")
sequence = sequence.translate(tab)[::-1]](https://image.slidesharecdn.com/2015bioinformaticspythonstringswimvancriekinge-151005212214-lva1-app6891/85/2015-bioinformatics-python_strings_wim_vancriekinge-53-320.jpg)