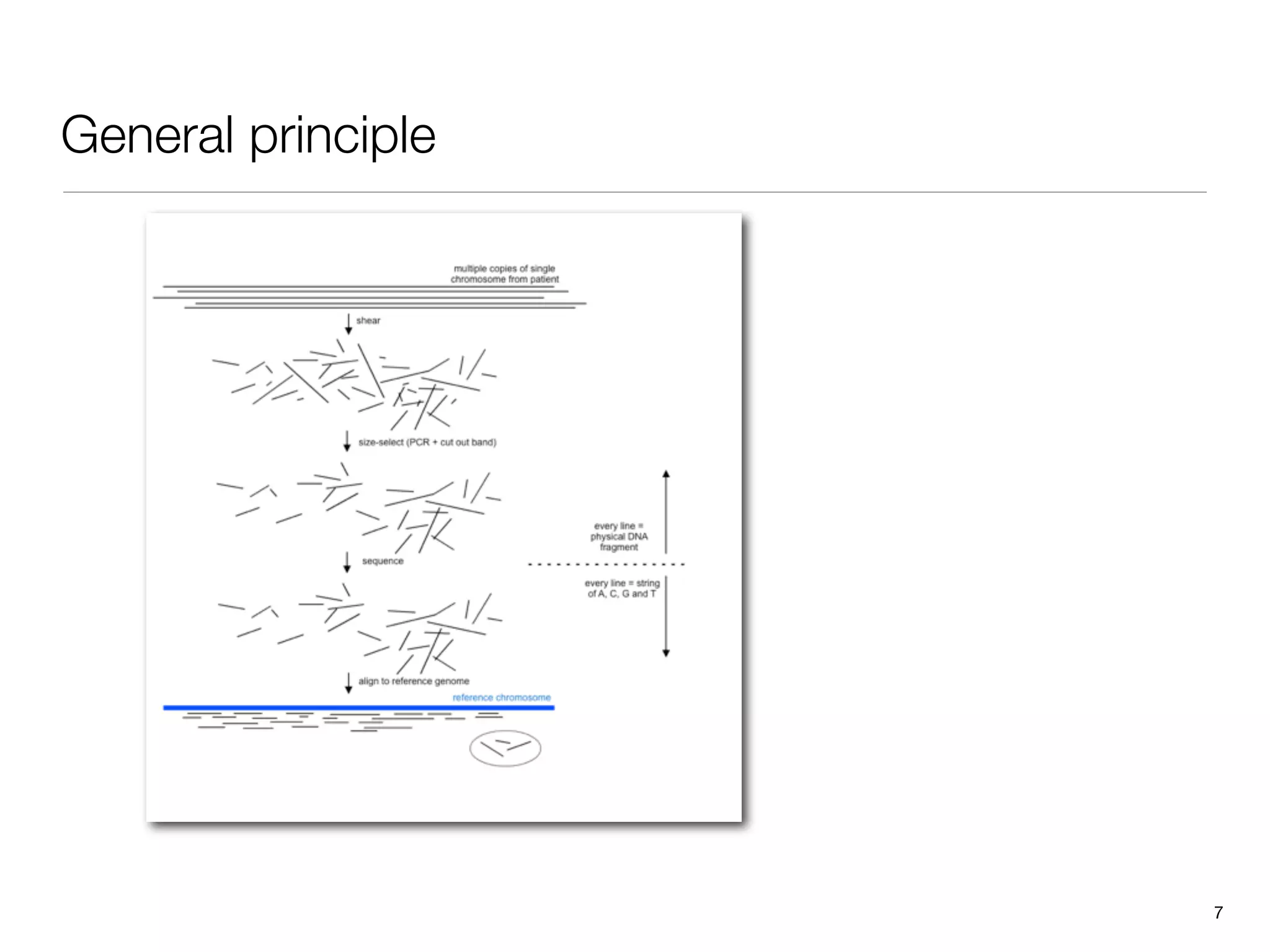
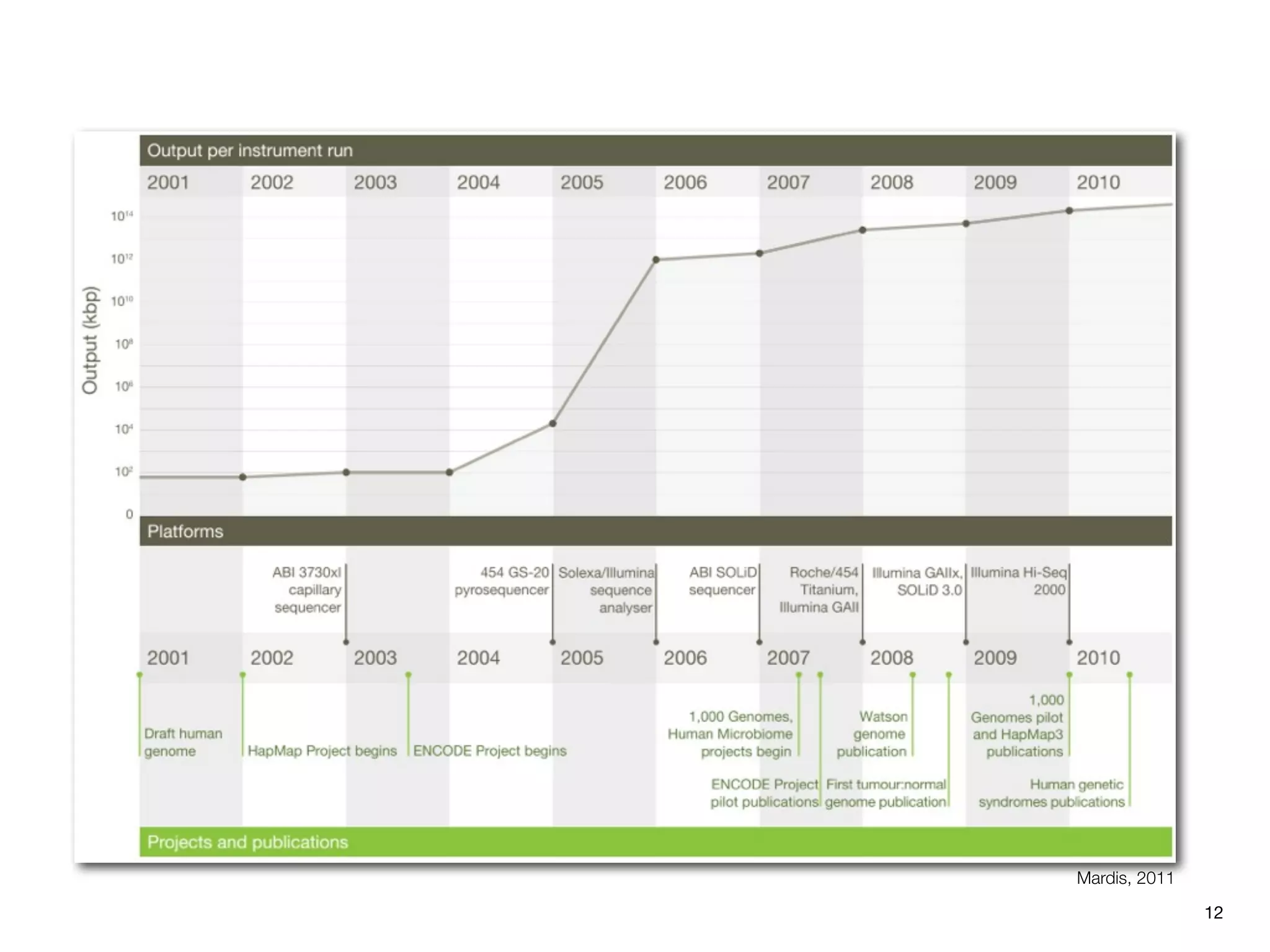
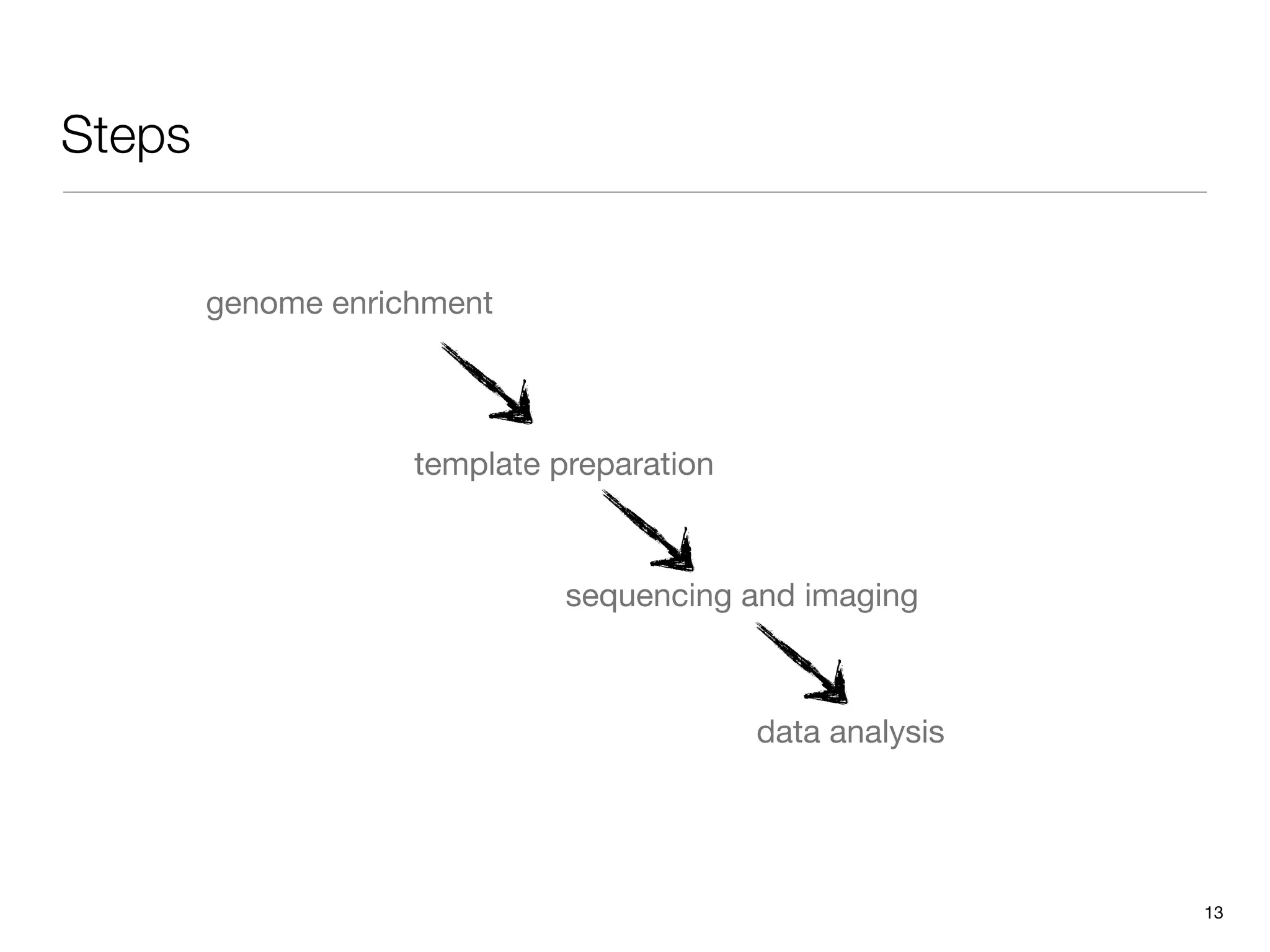
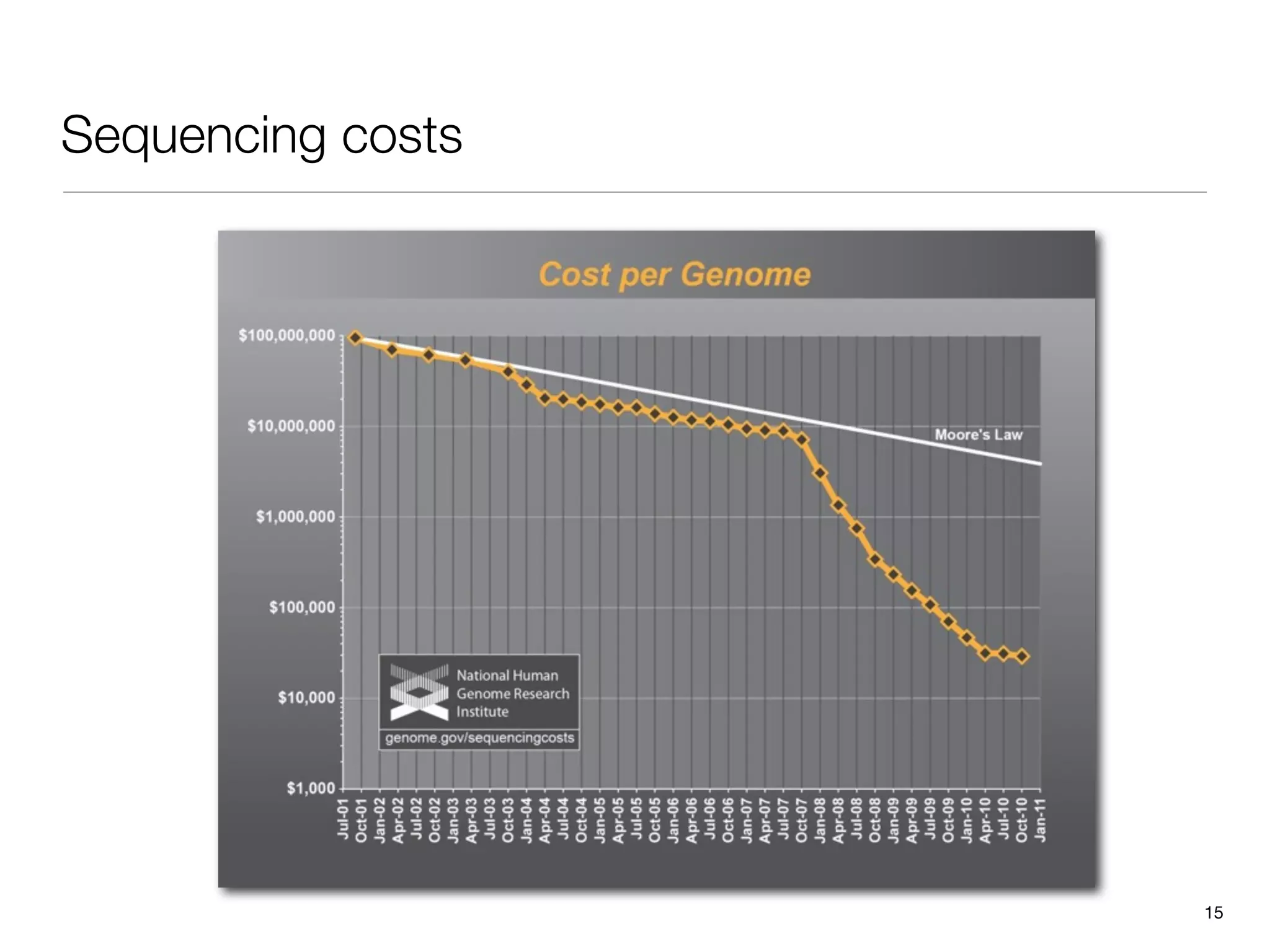
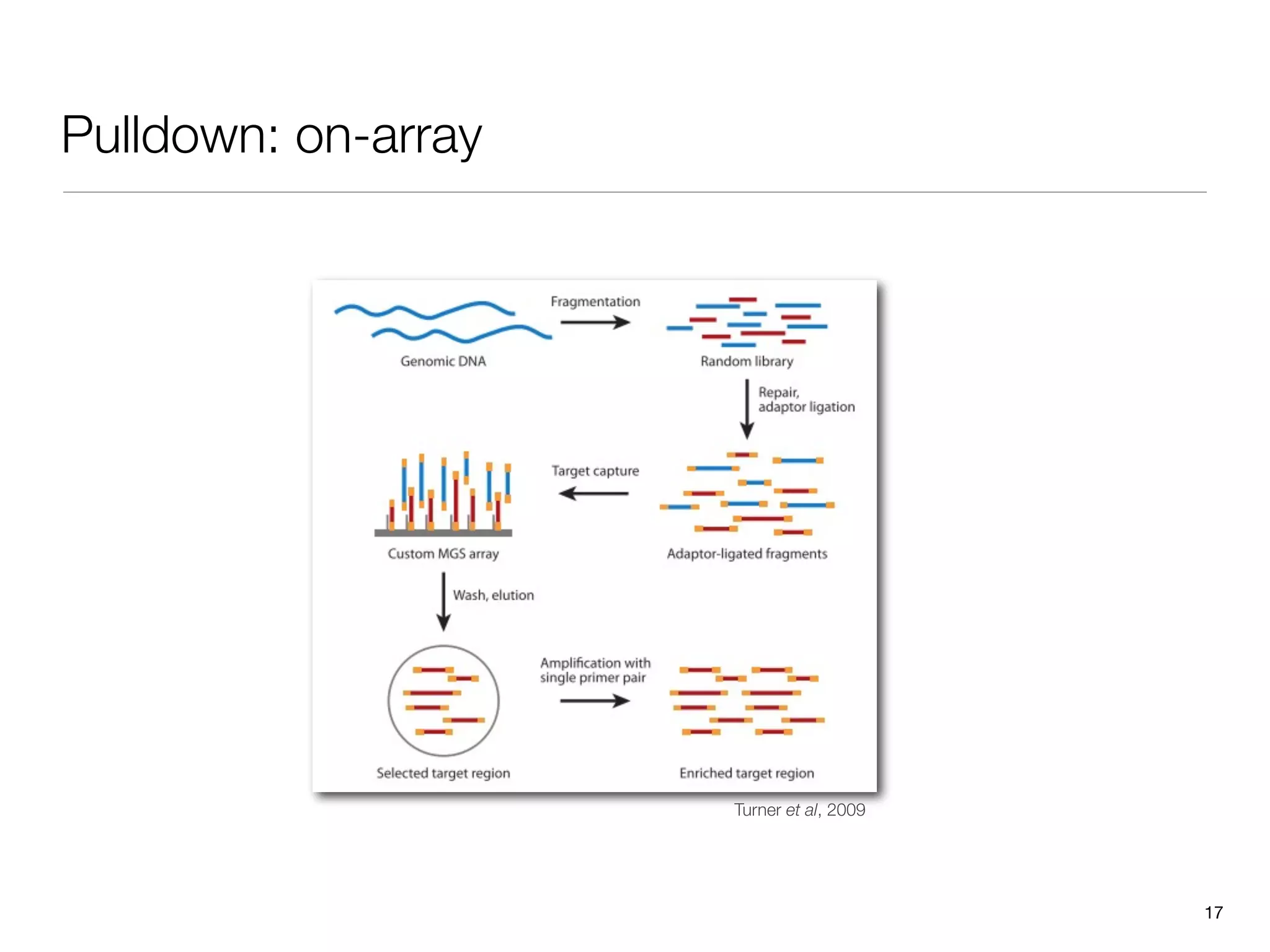
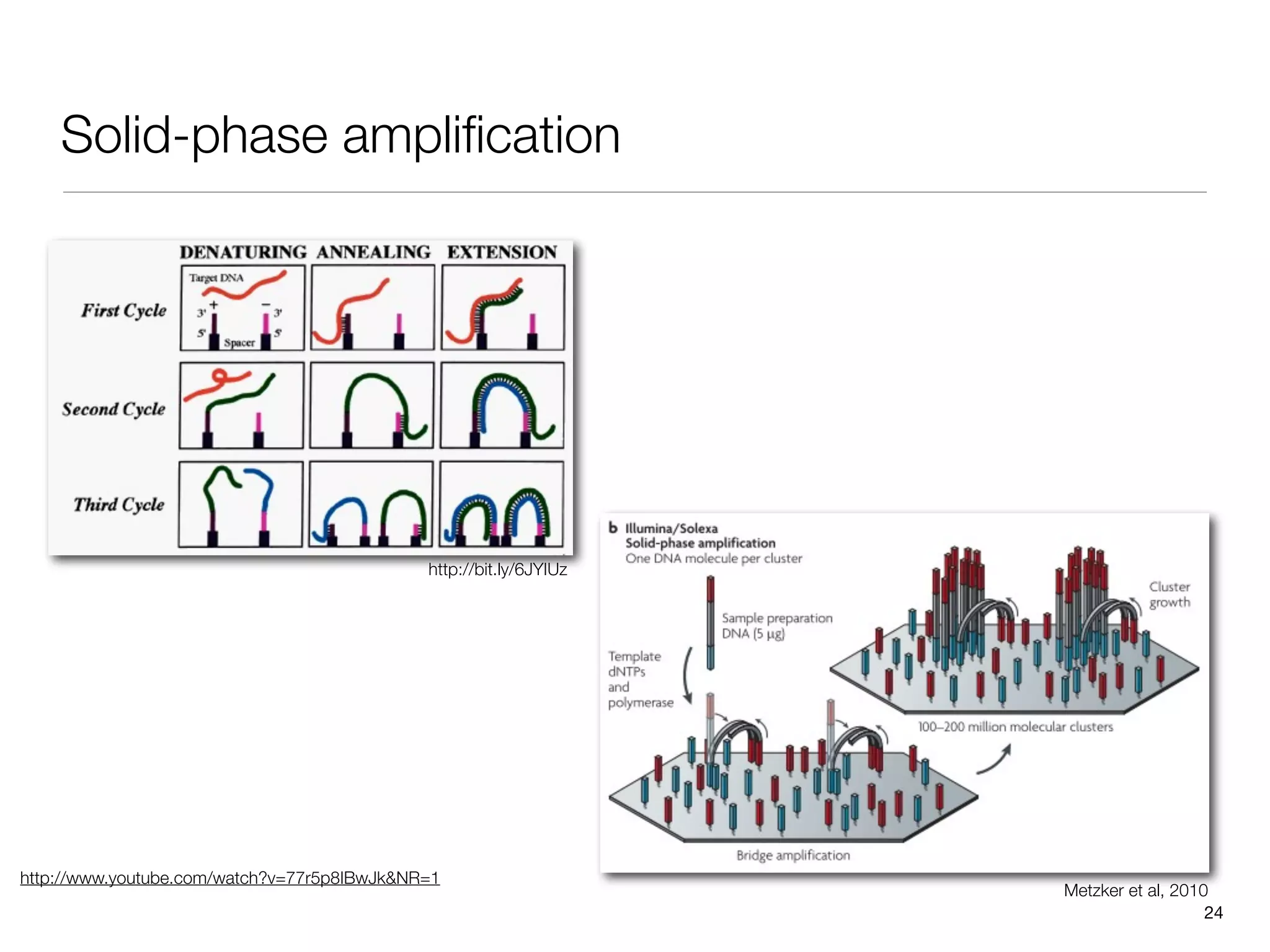
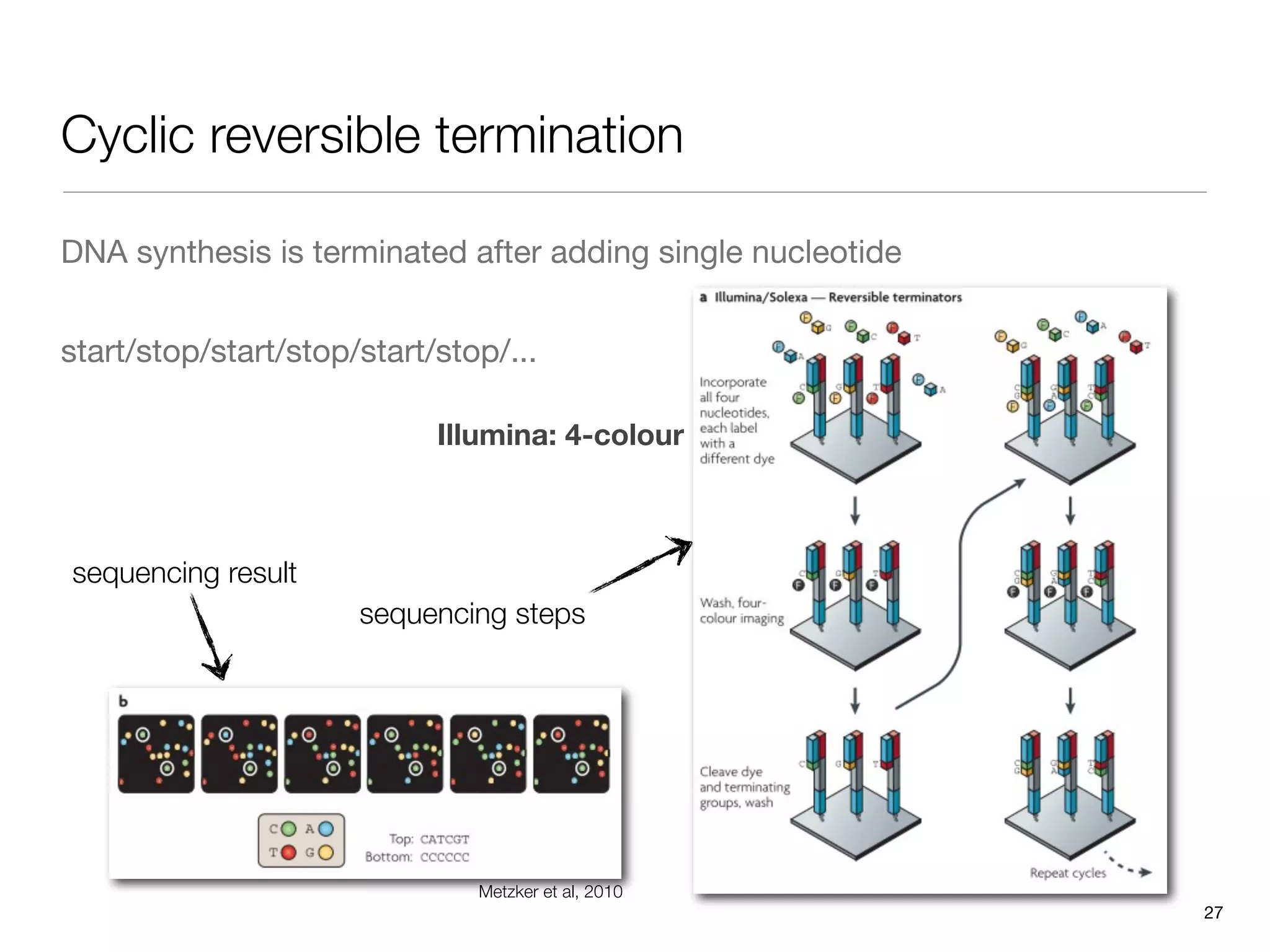
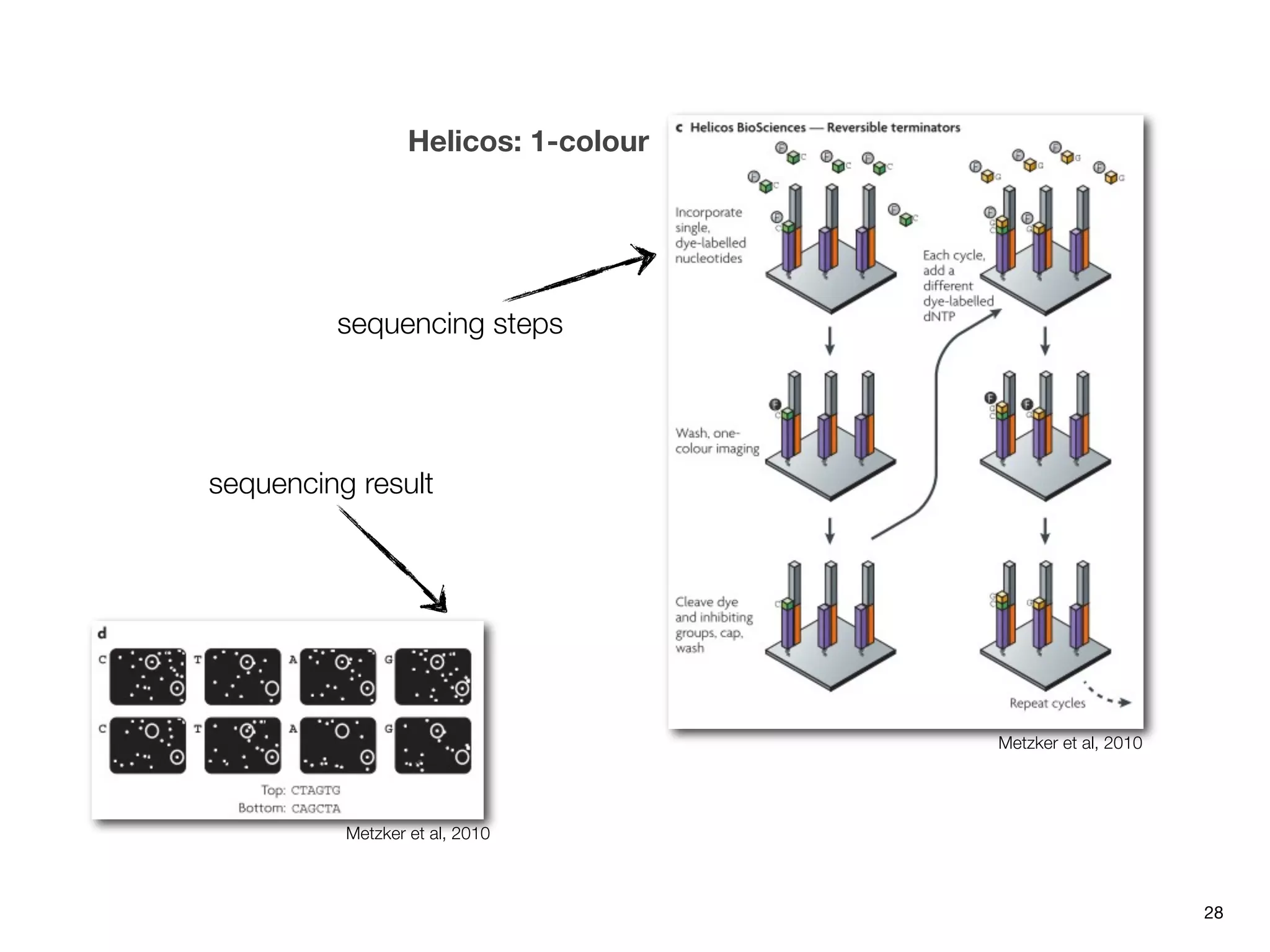
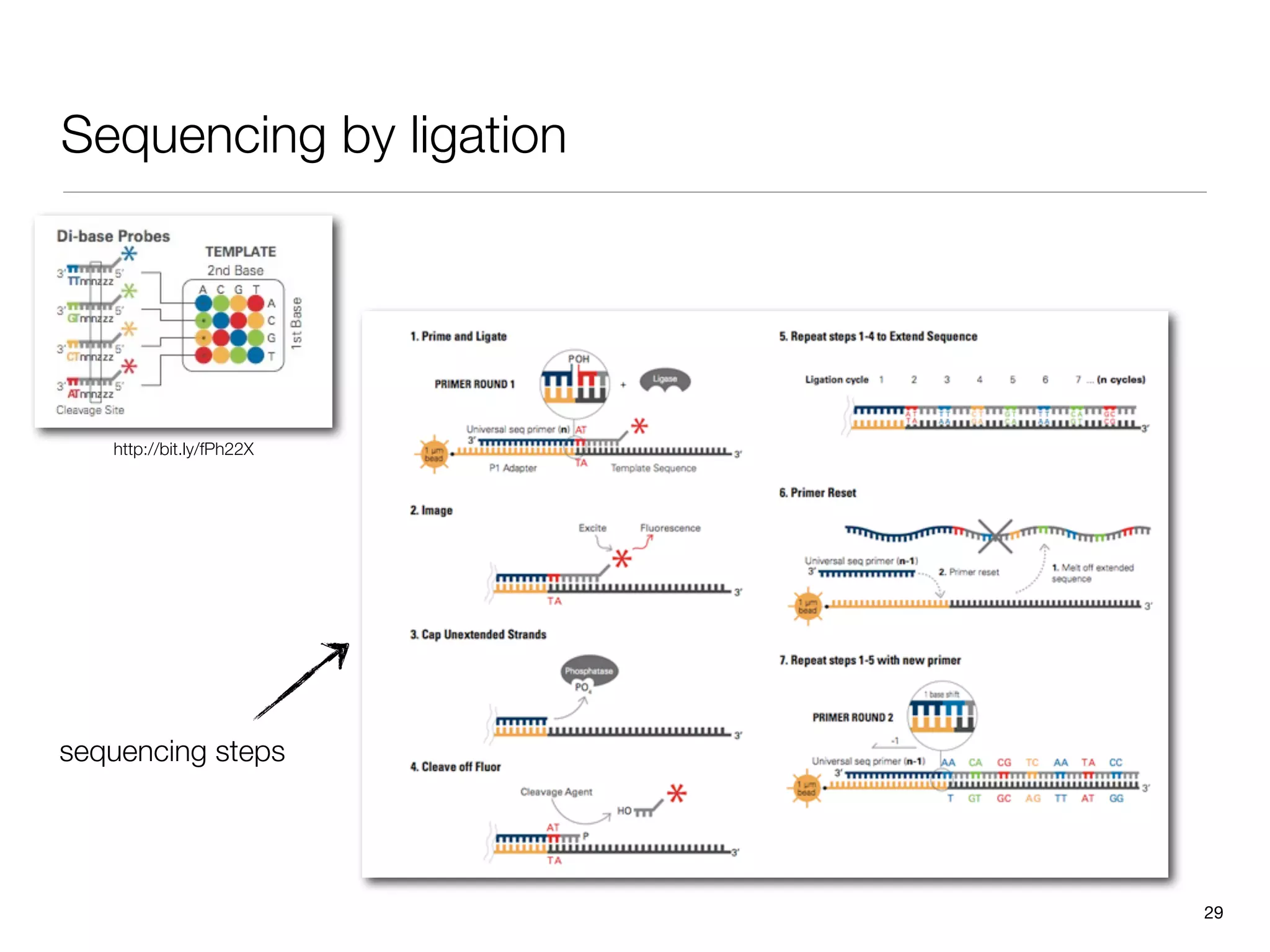
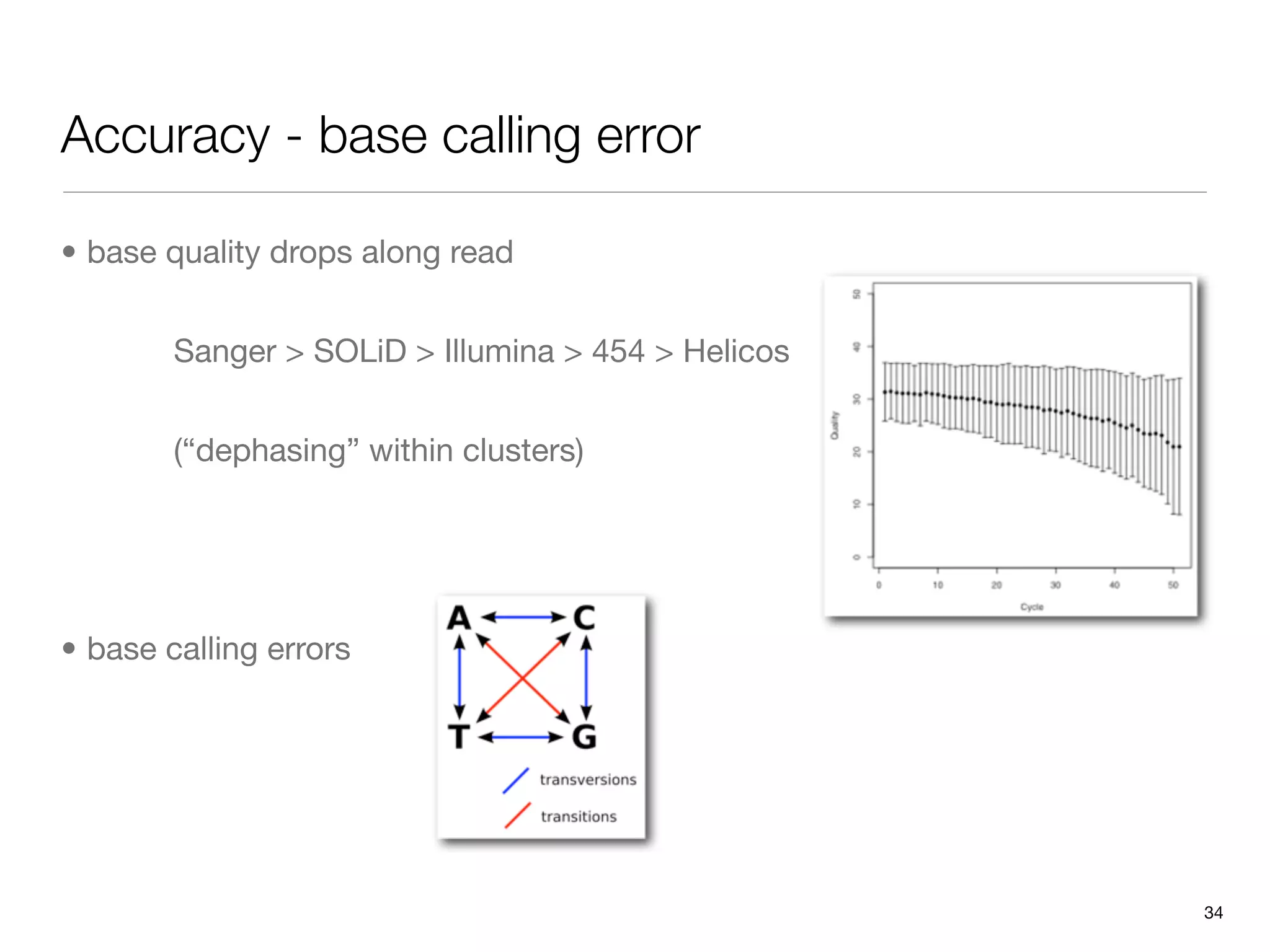
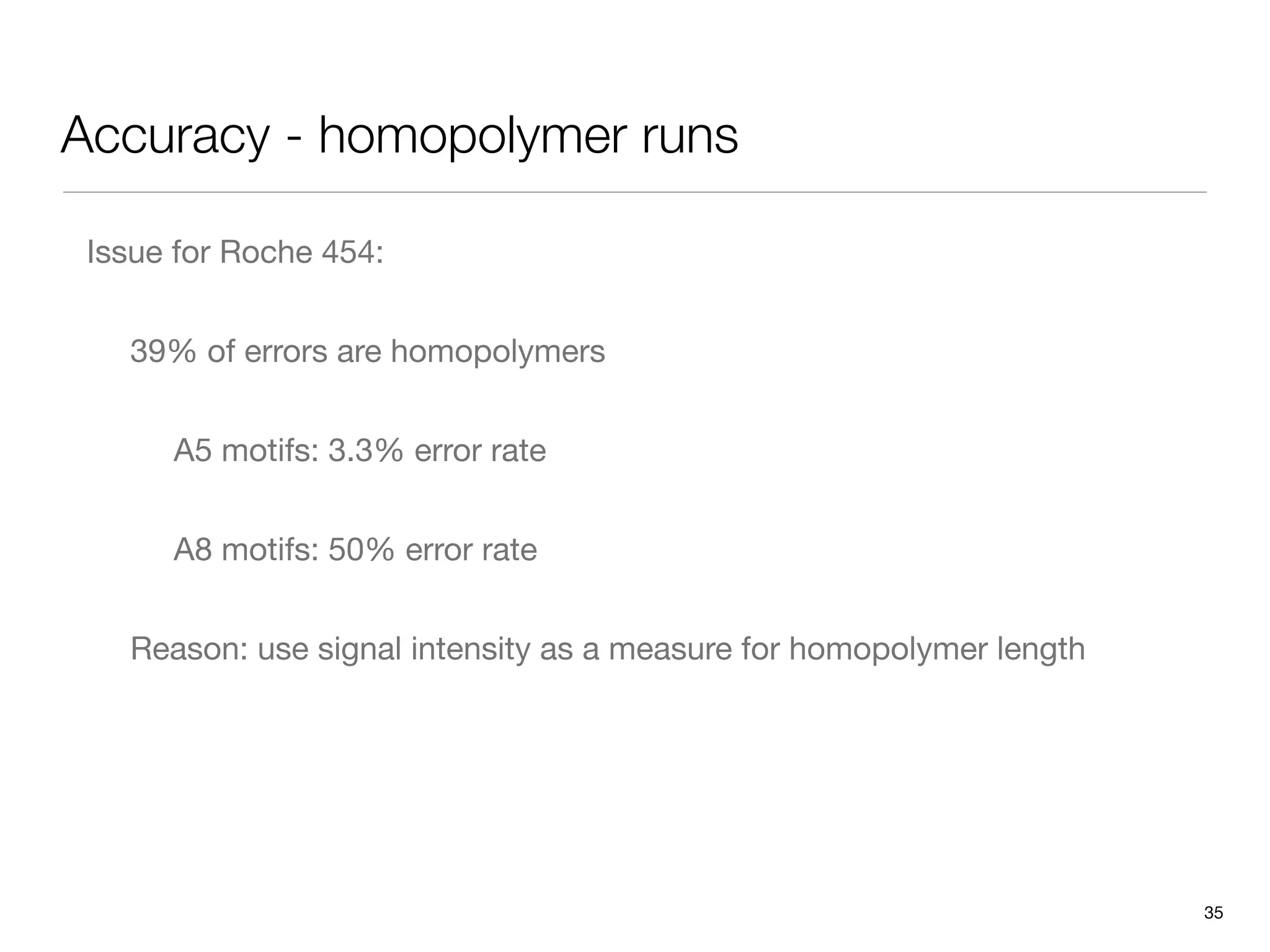
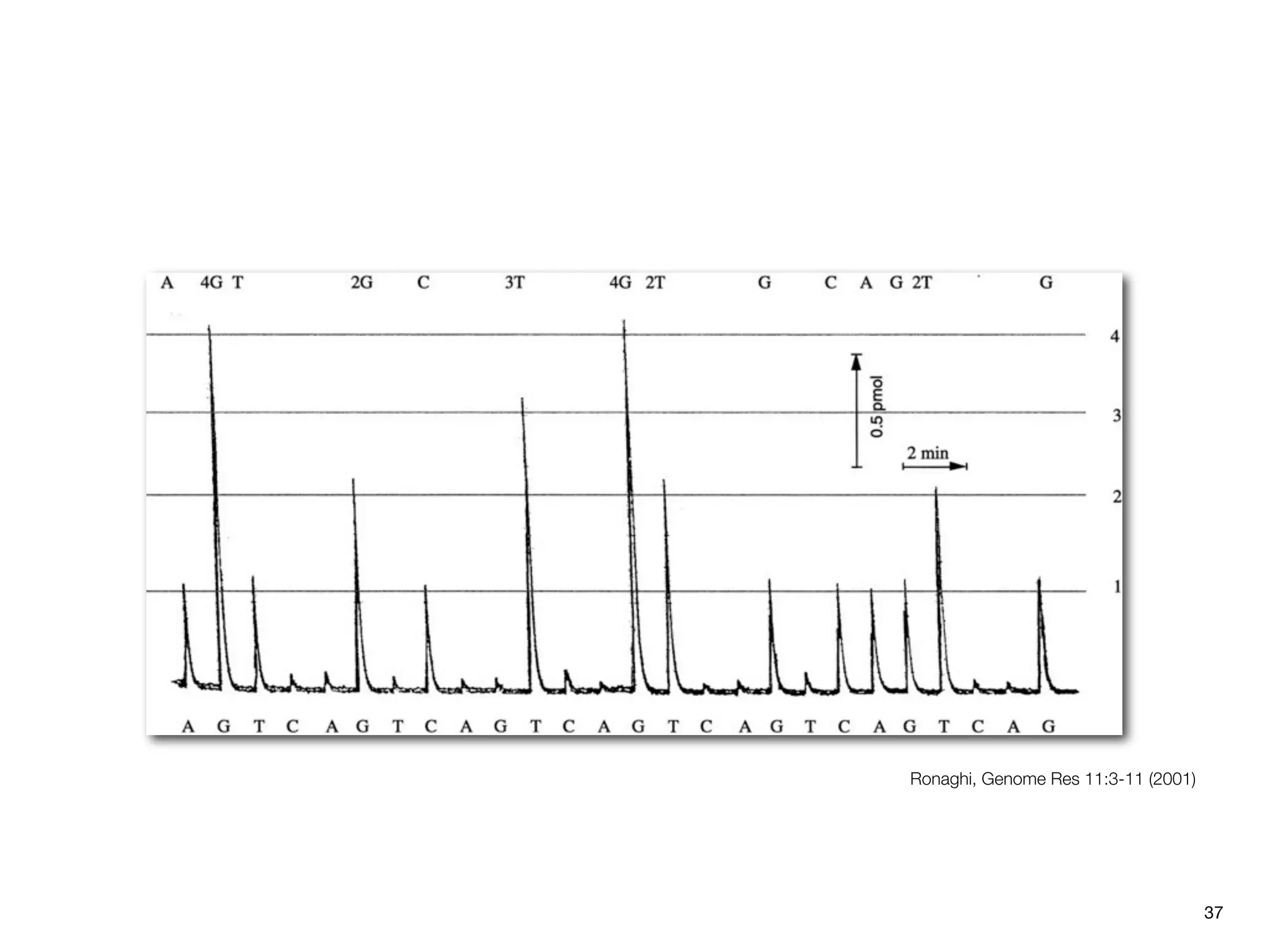
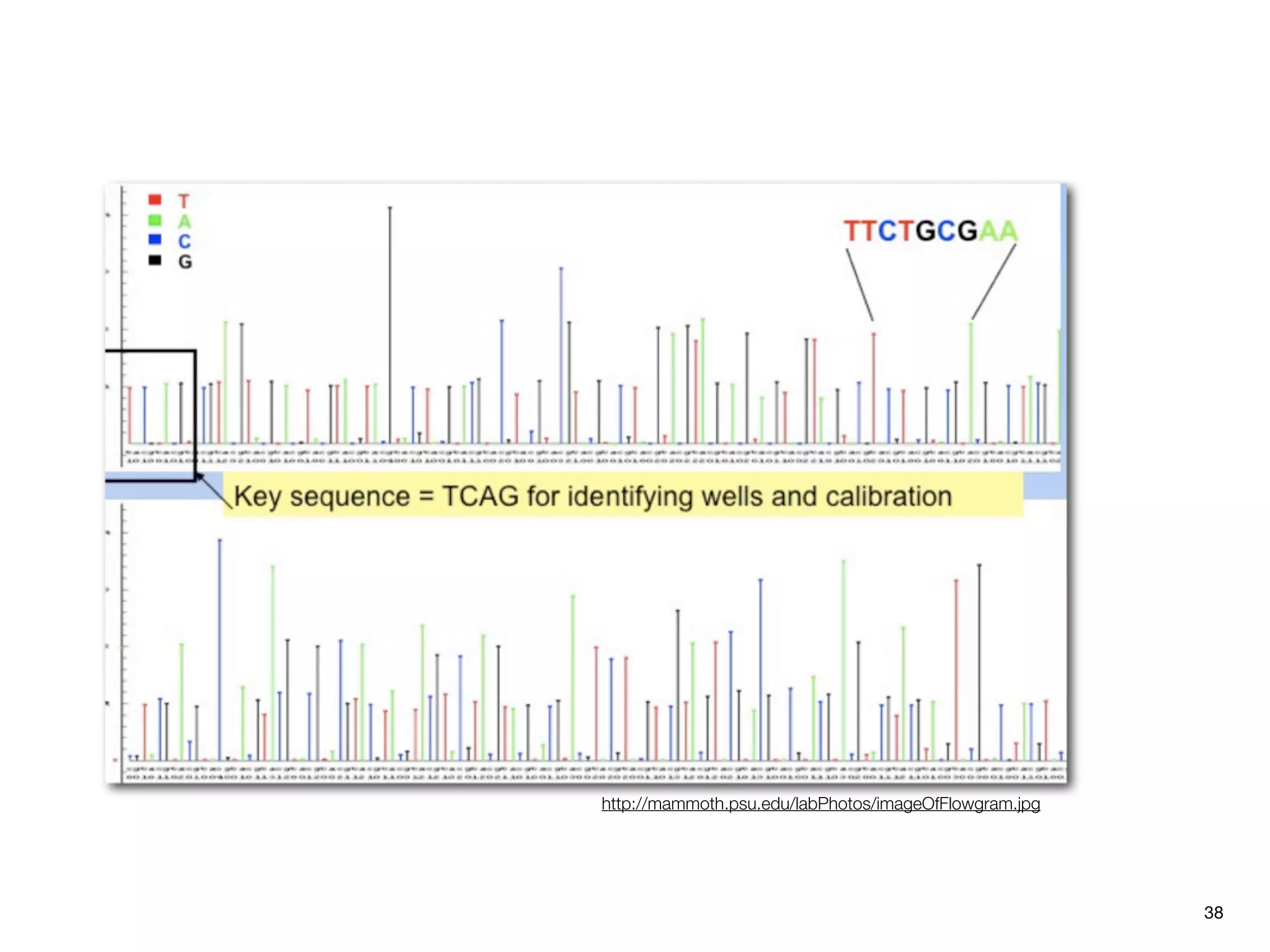
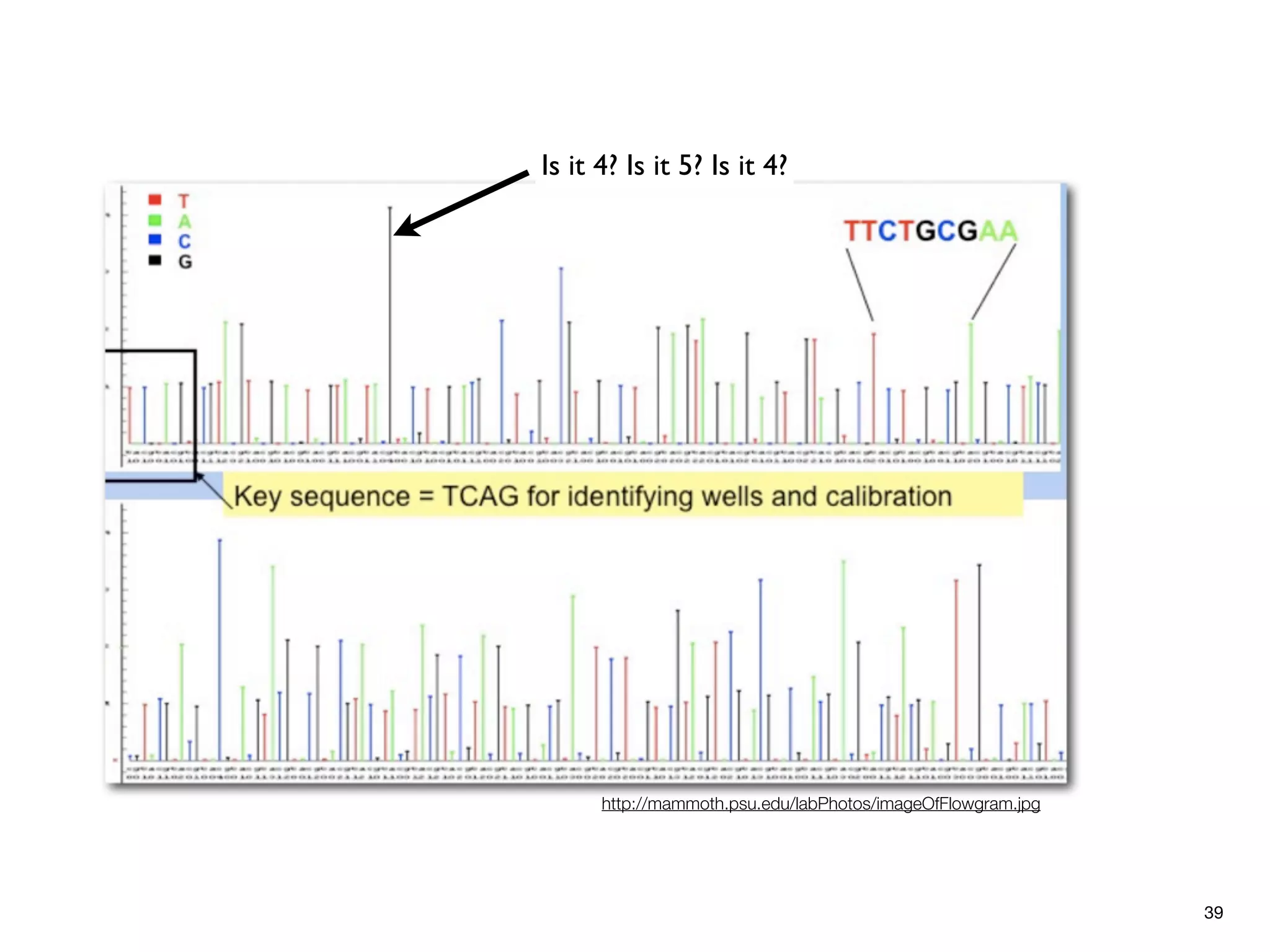
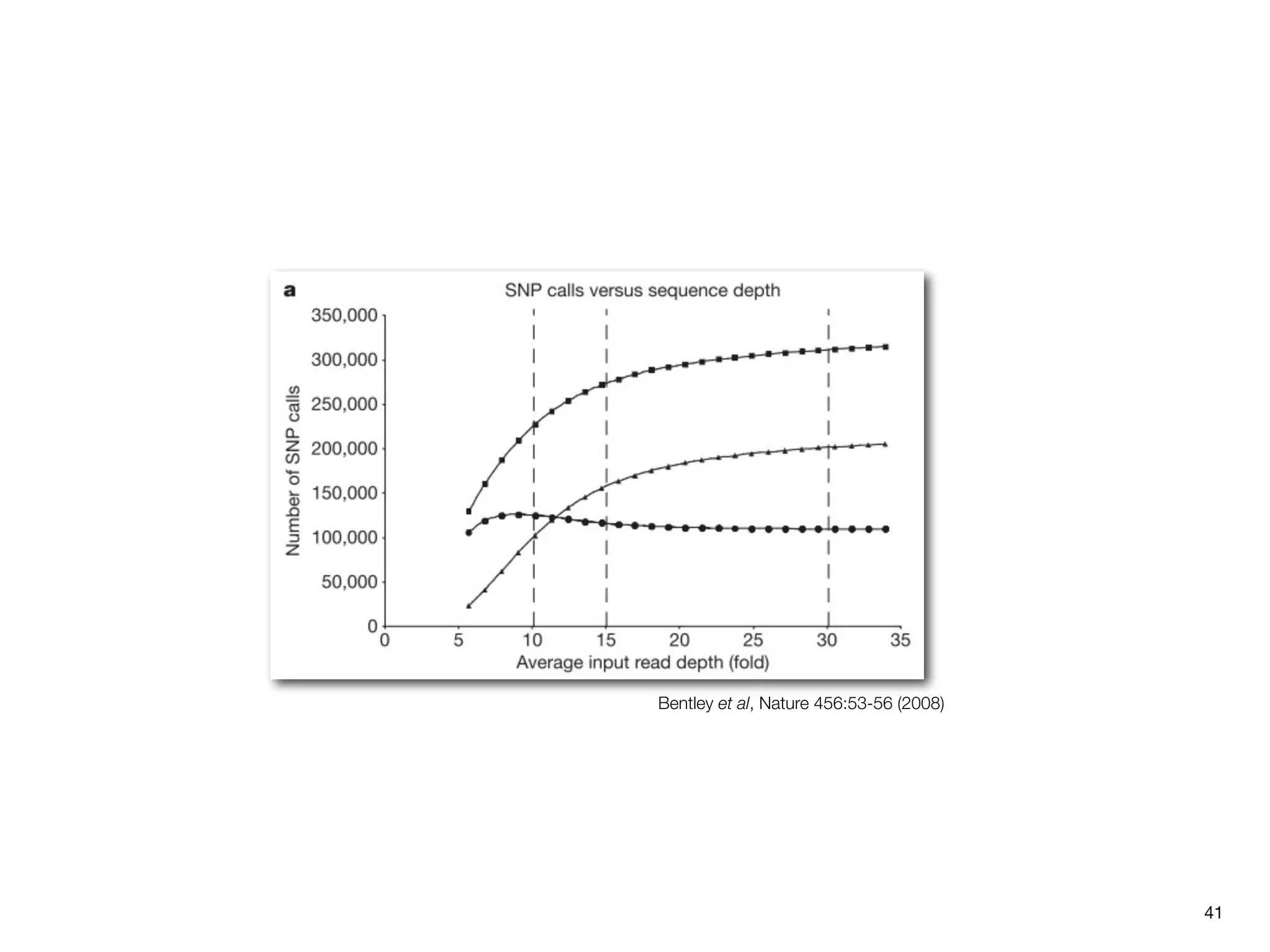
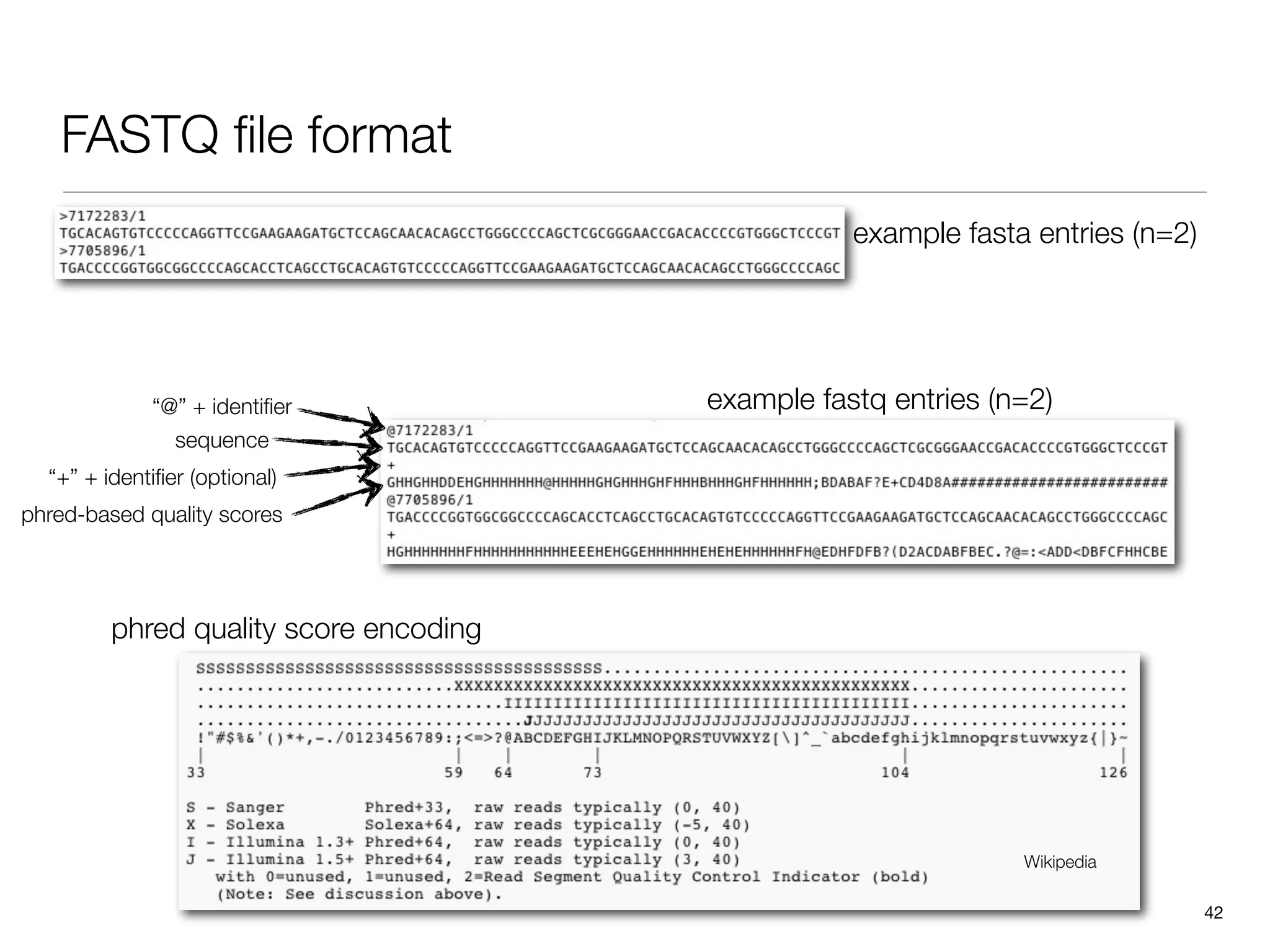
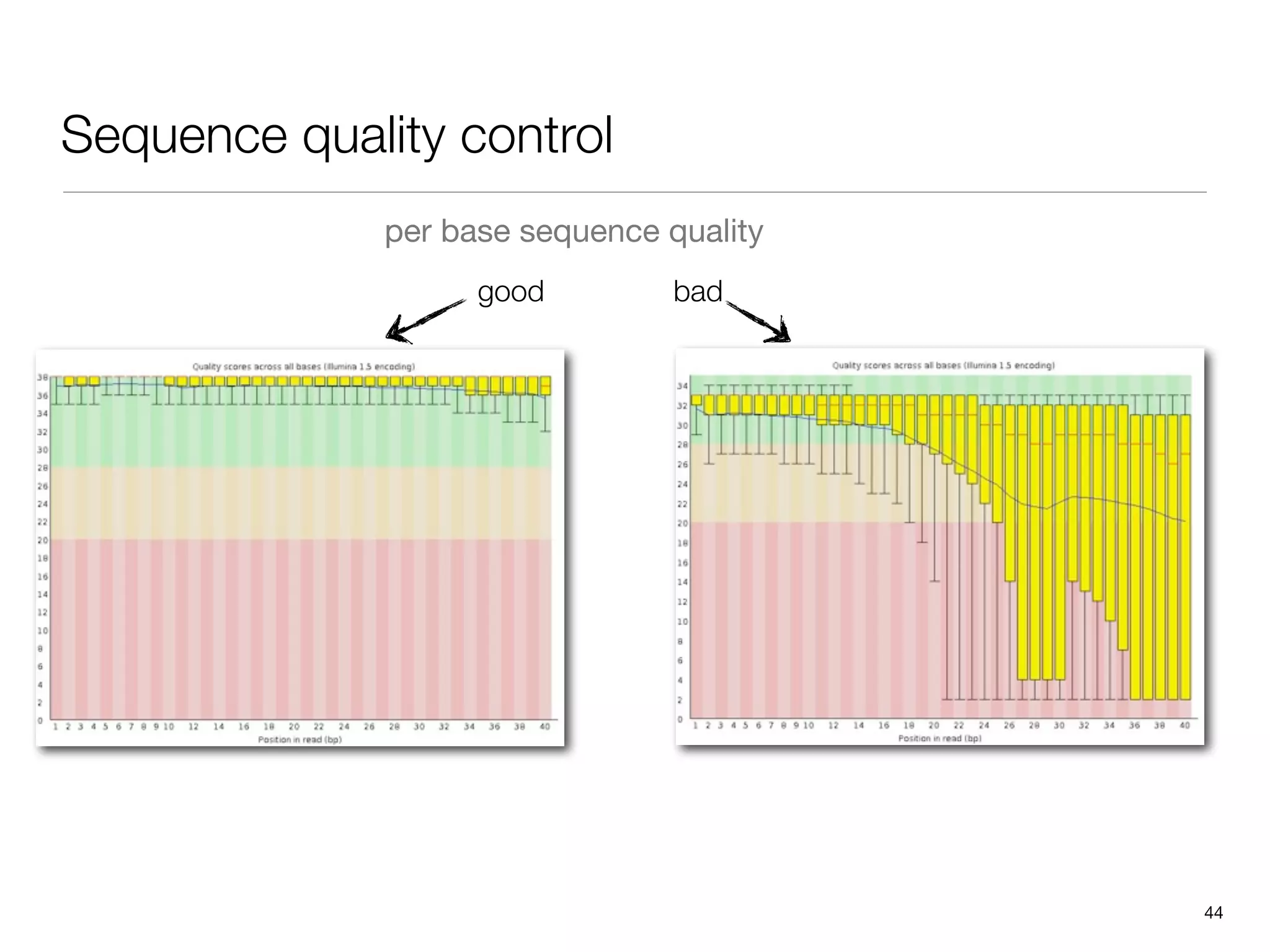
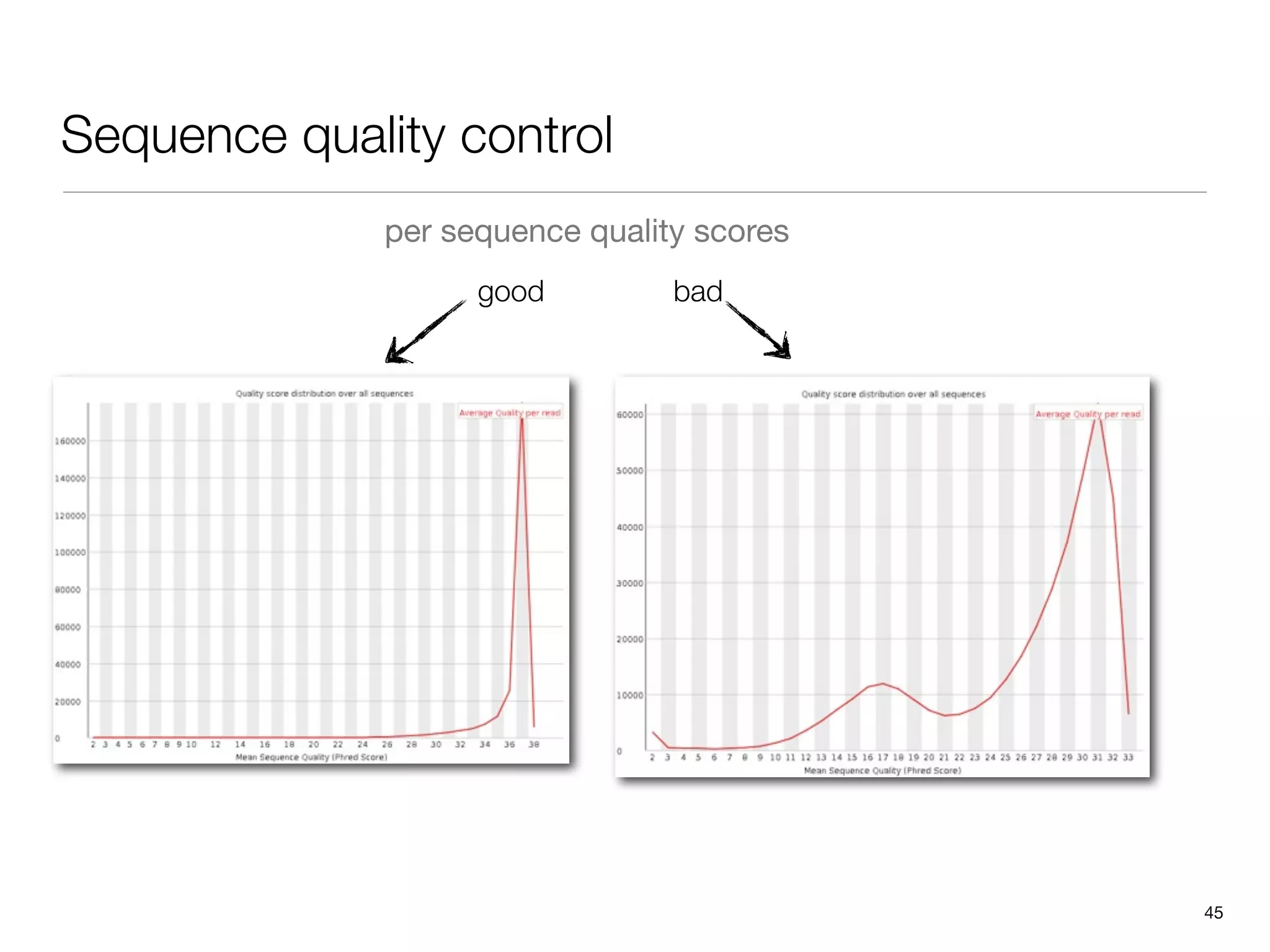
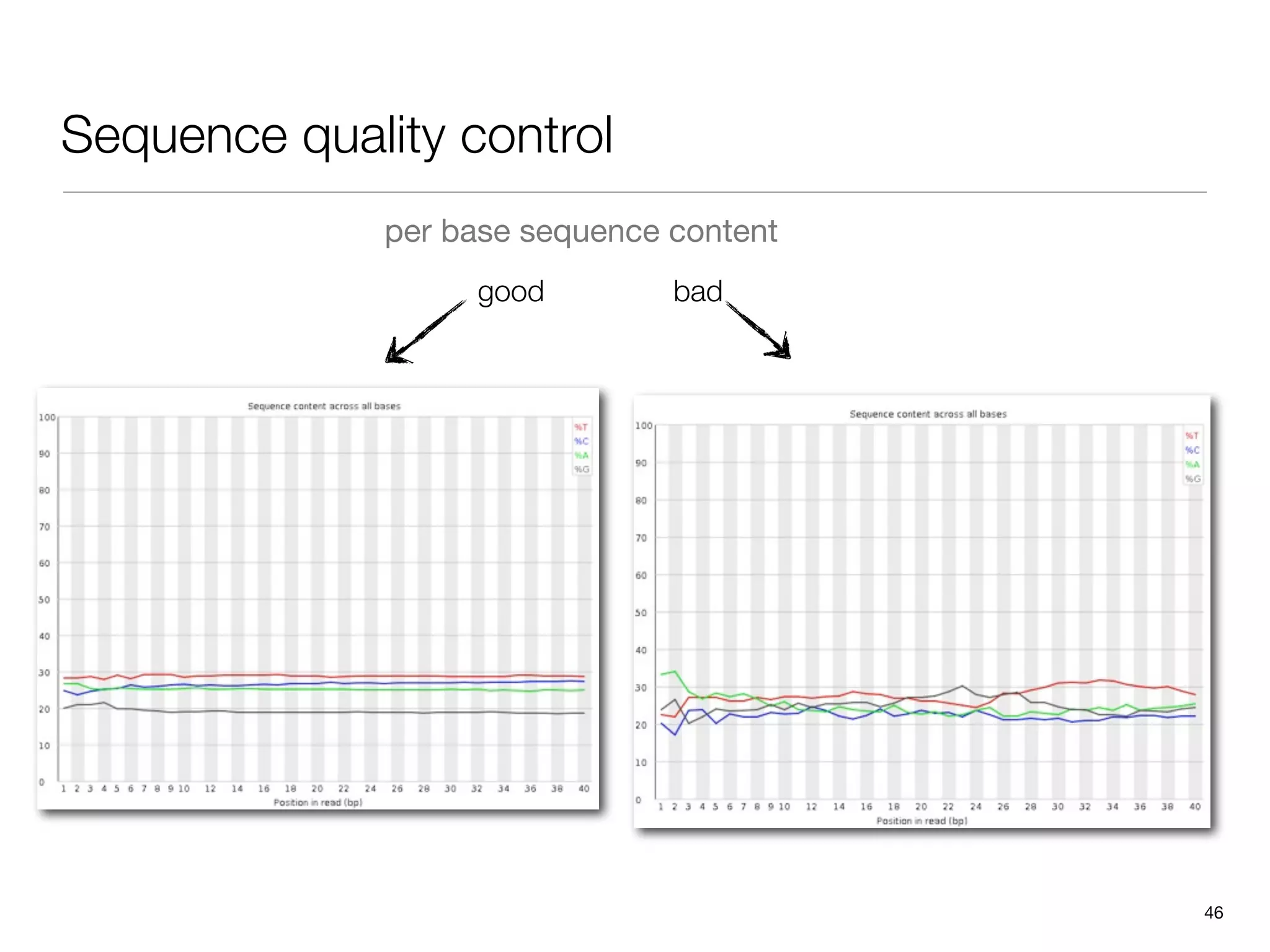
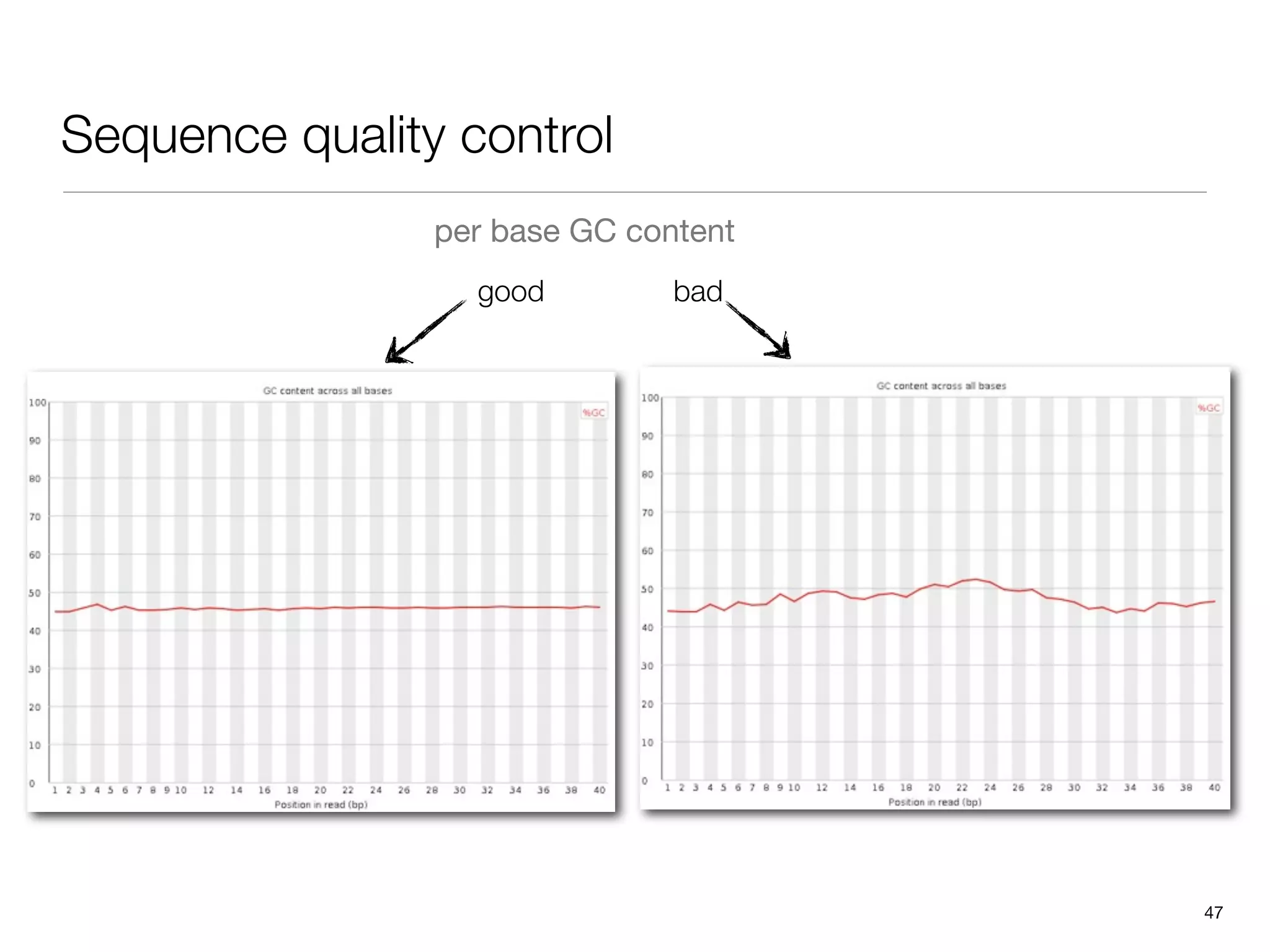
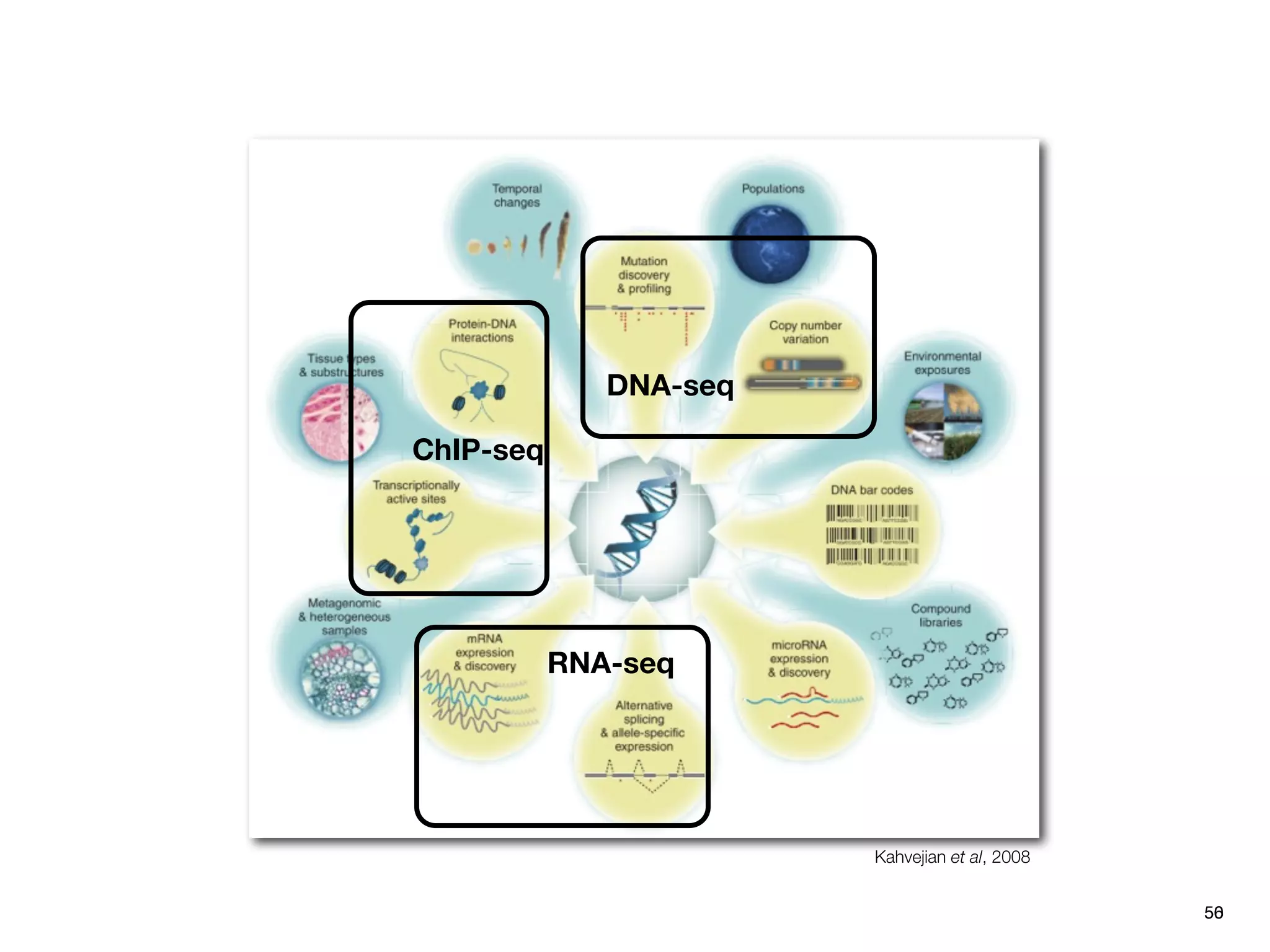
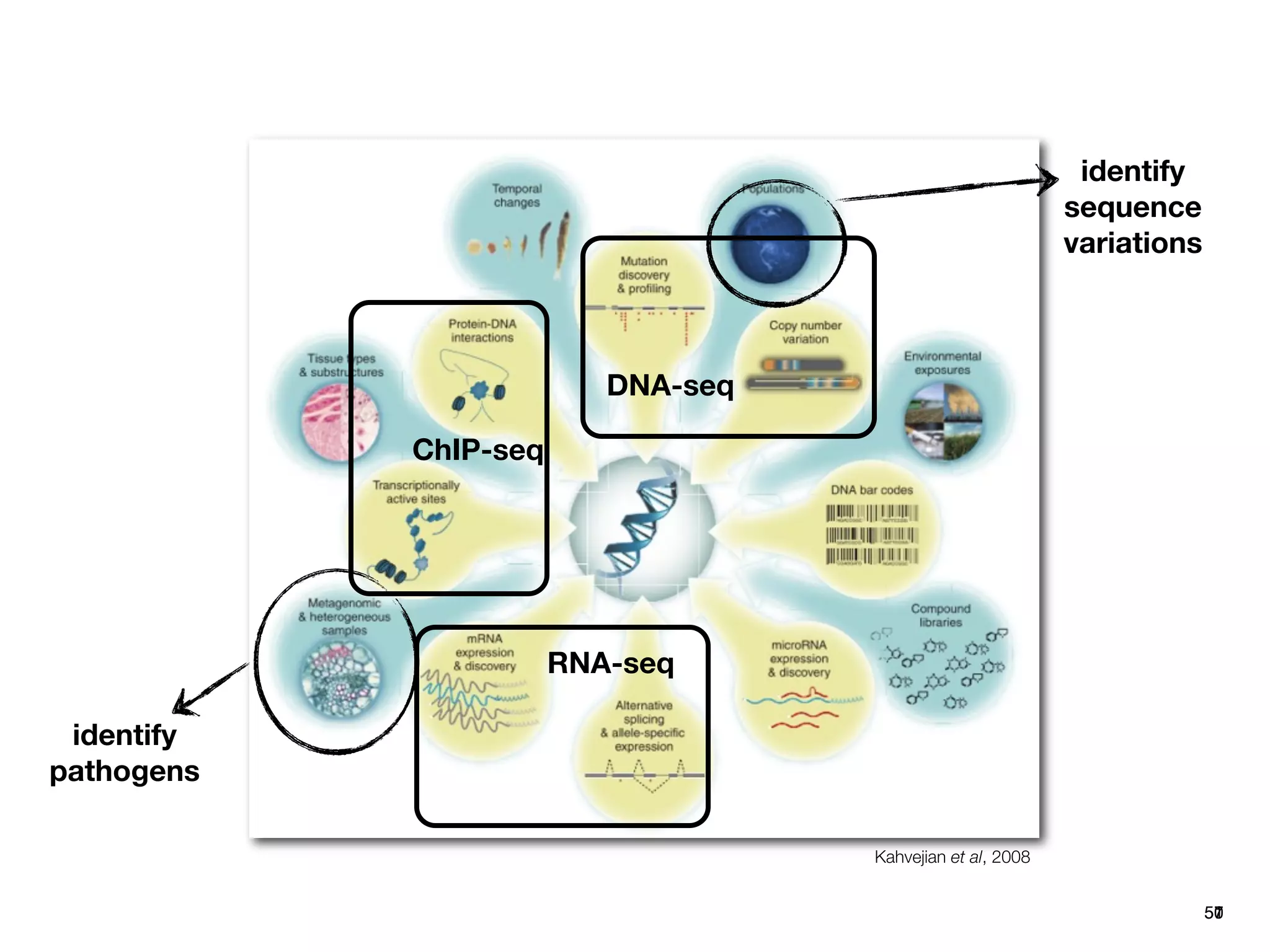
This document provides an overview of next-generation sequencing technologies and their applications. It discusses genome enrichment techniques to isolate targeted regions for sequencing. It also describes template preparation methods like emulsion PCR and solid-phase amplification. Finally, it reviews various sequencing platforms like Illumina, SOLiD, 454 and details the sequencing and imaging processes. There are exercises proposed to work with sequencing data files in Galaxy.
![[I0D51A] Bioinformatics: High-Throughput Analysis
Next-generation sequencing. Part 1: Technologies
Prof Jan Aerts
Faculty of Engineering - ESAT/SCD
jan.aerts@esat.kuleuven.be
TA: Alejandro Sifrim (alejandro.sifrim@esat.kuleuven.be)
1](https://image.slidesharecdn.com/i0d51a20110506-110505140616-phpapp02/75/Next-generation-sequencing-course-part-1-technologies-1-2048.jpg)