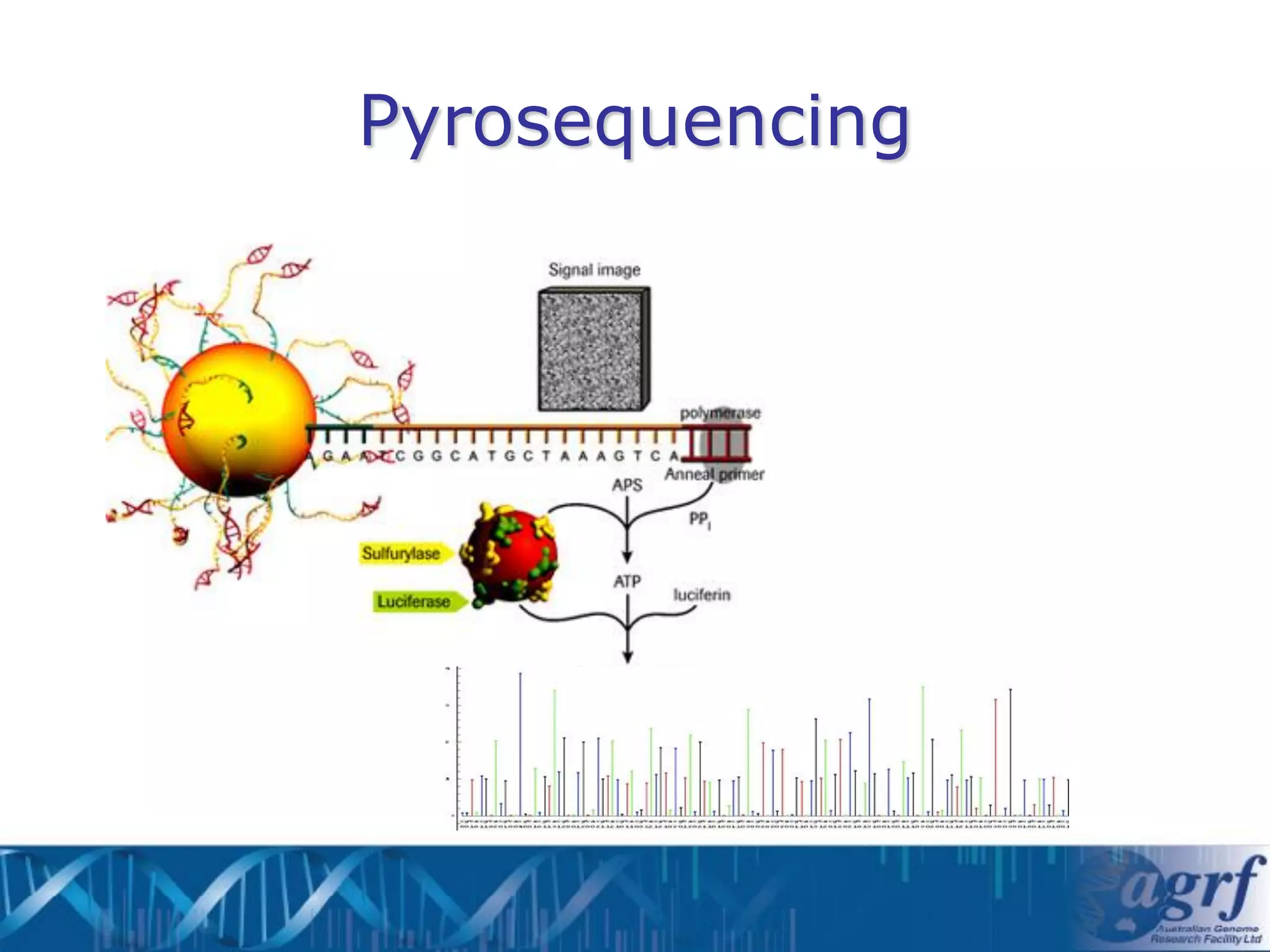
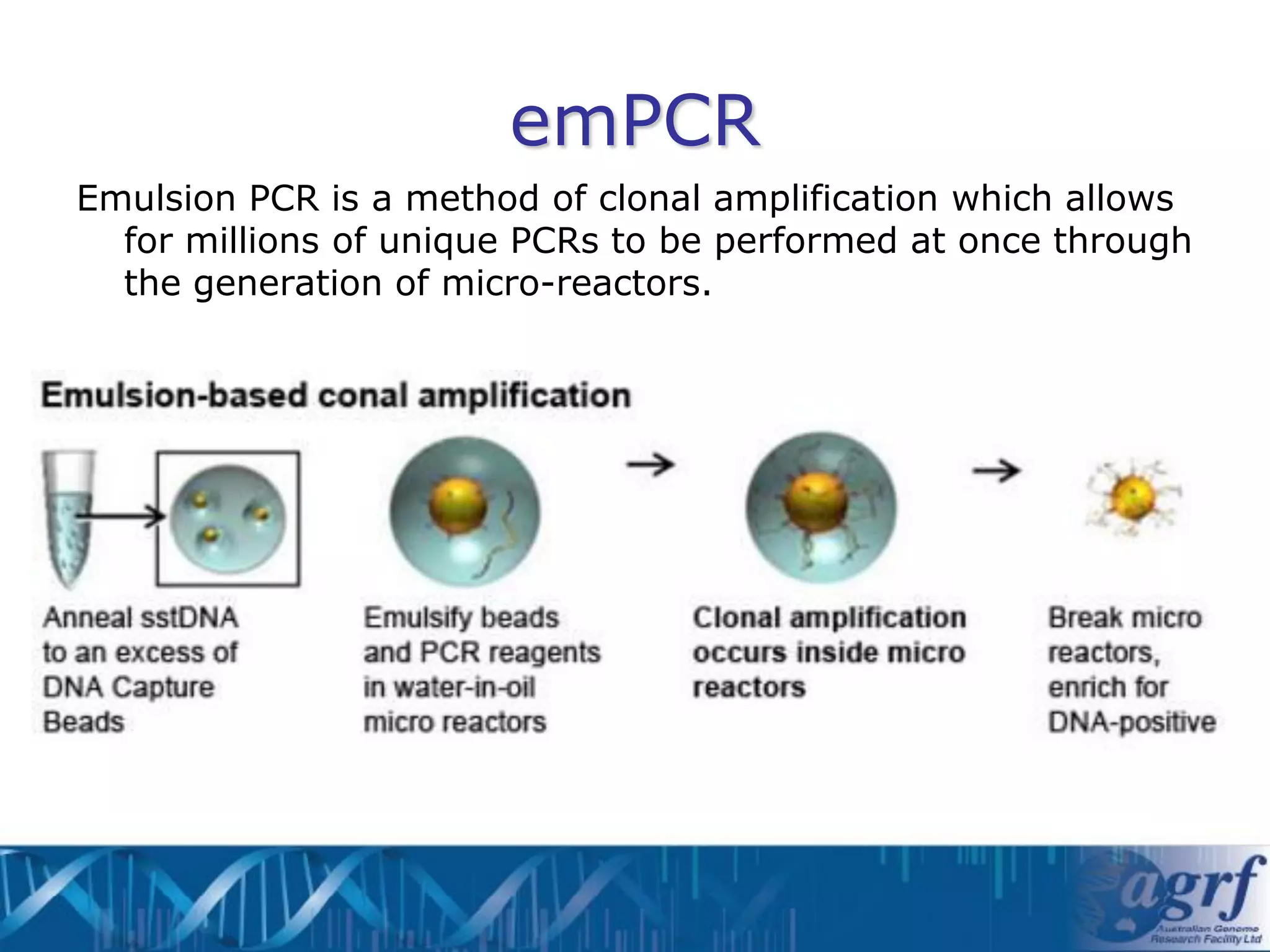
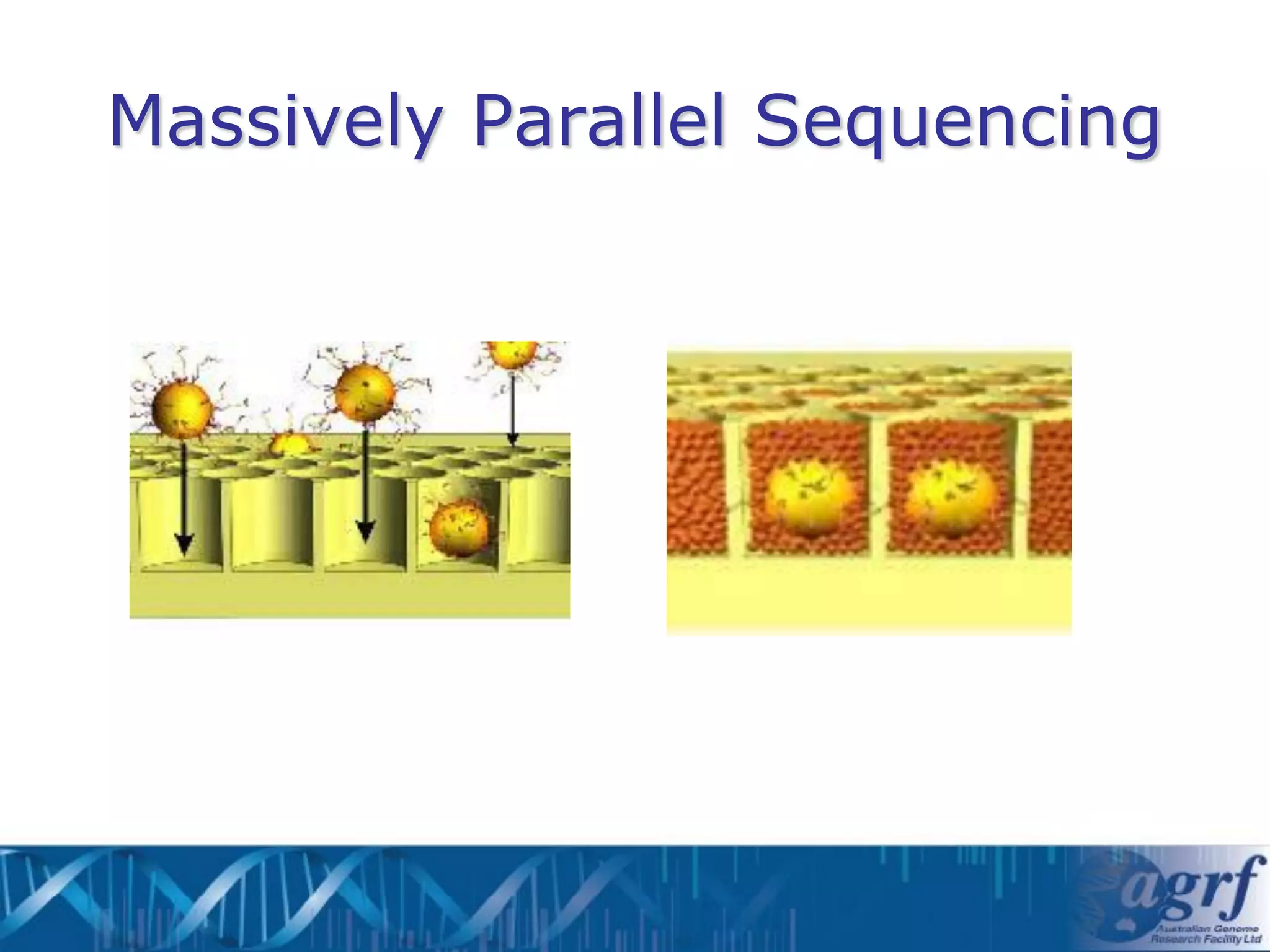
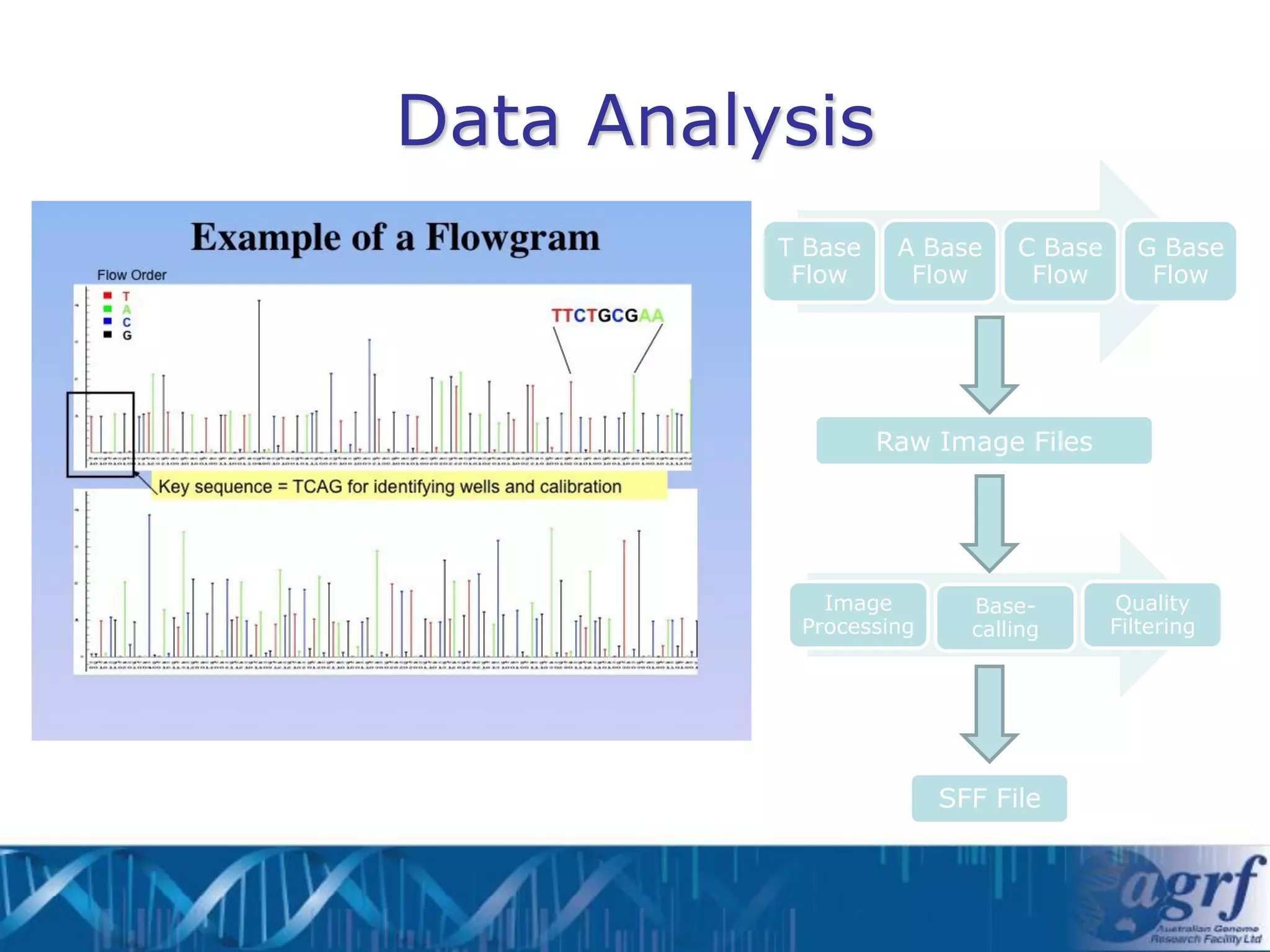
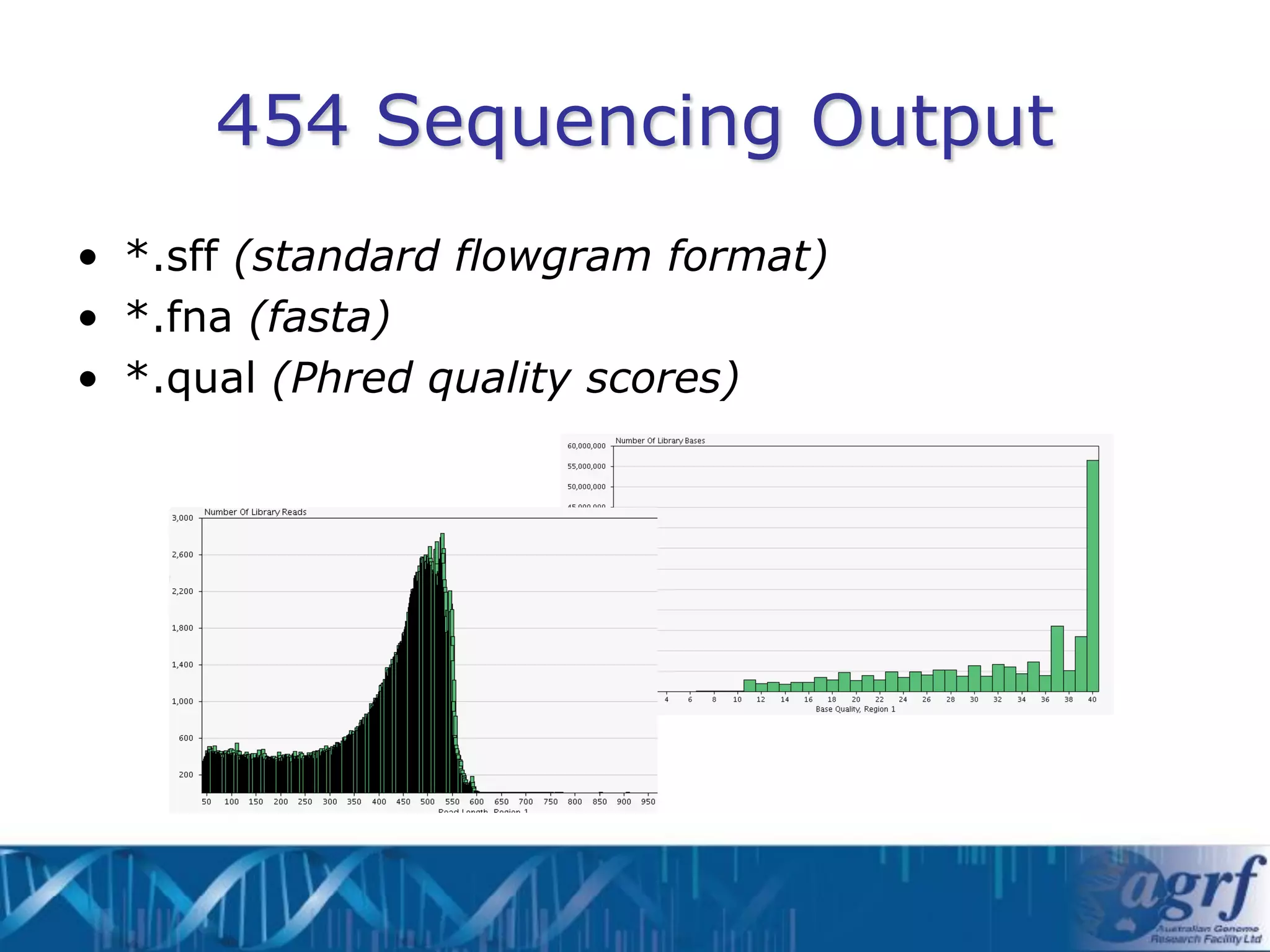
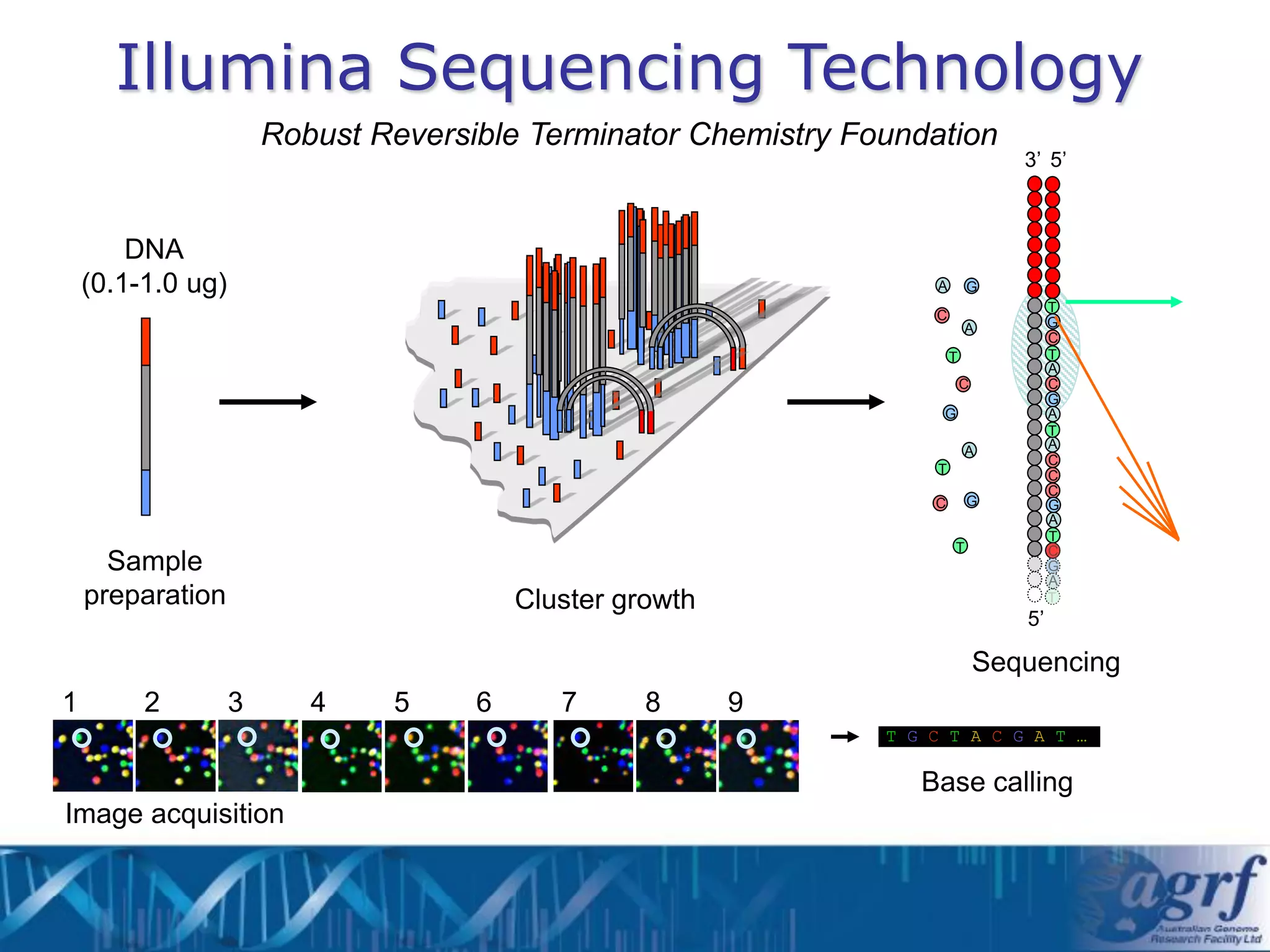
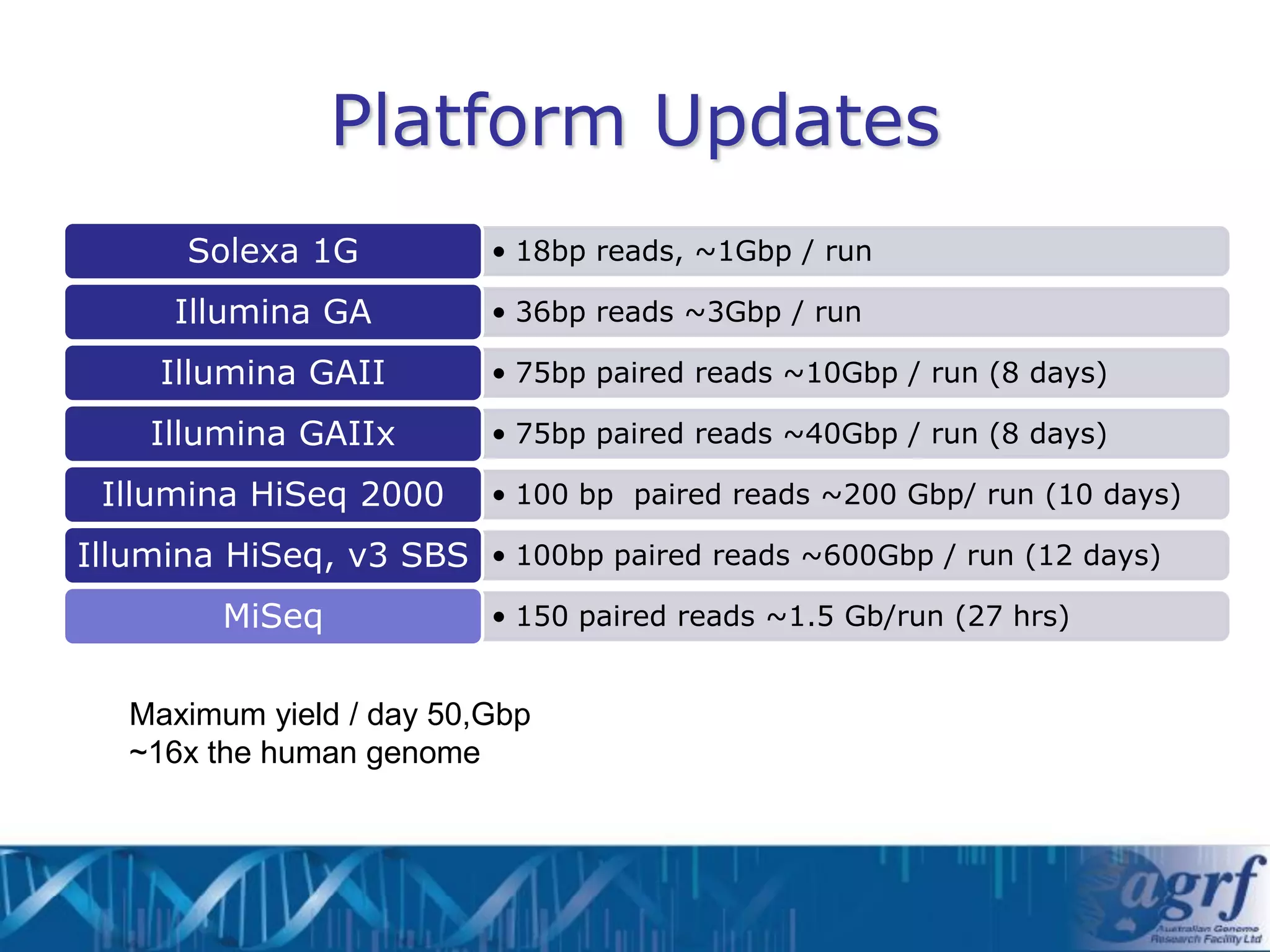
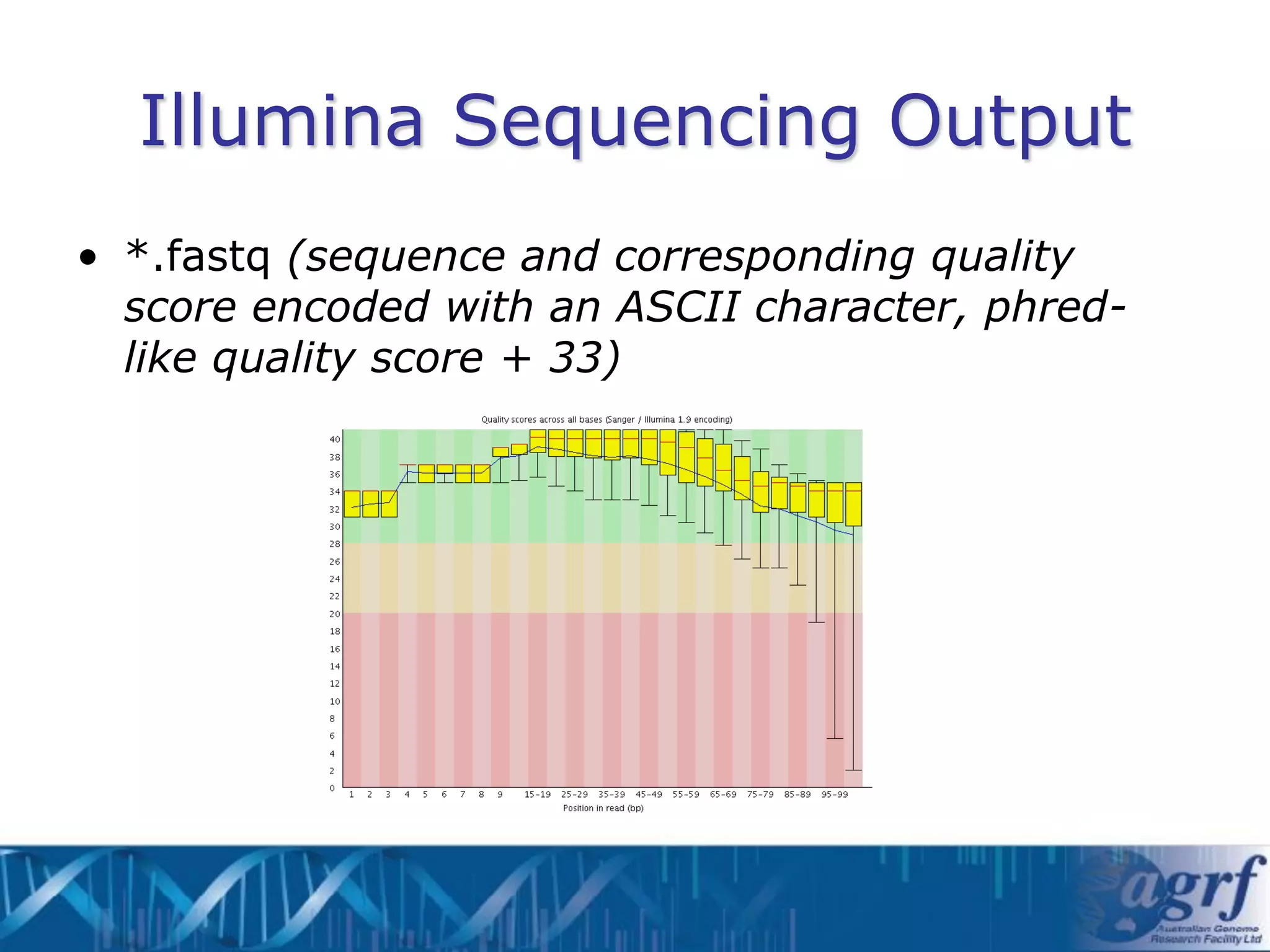
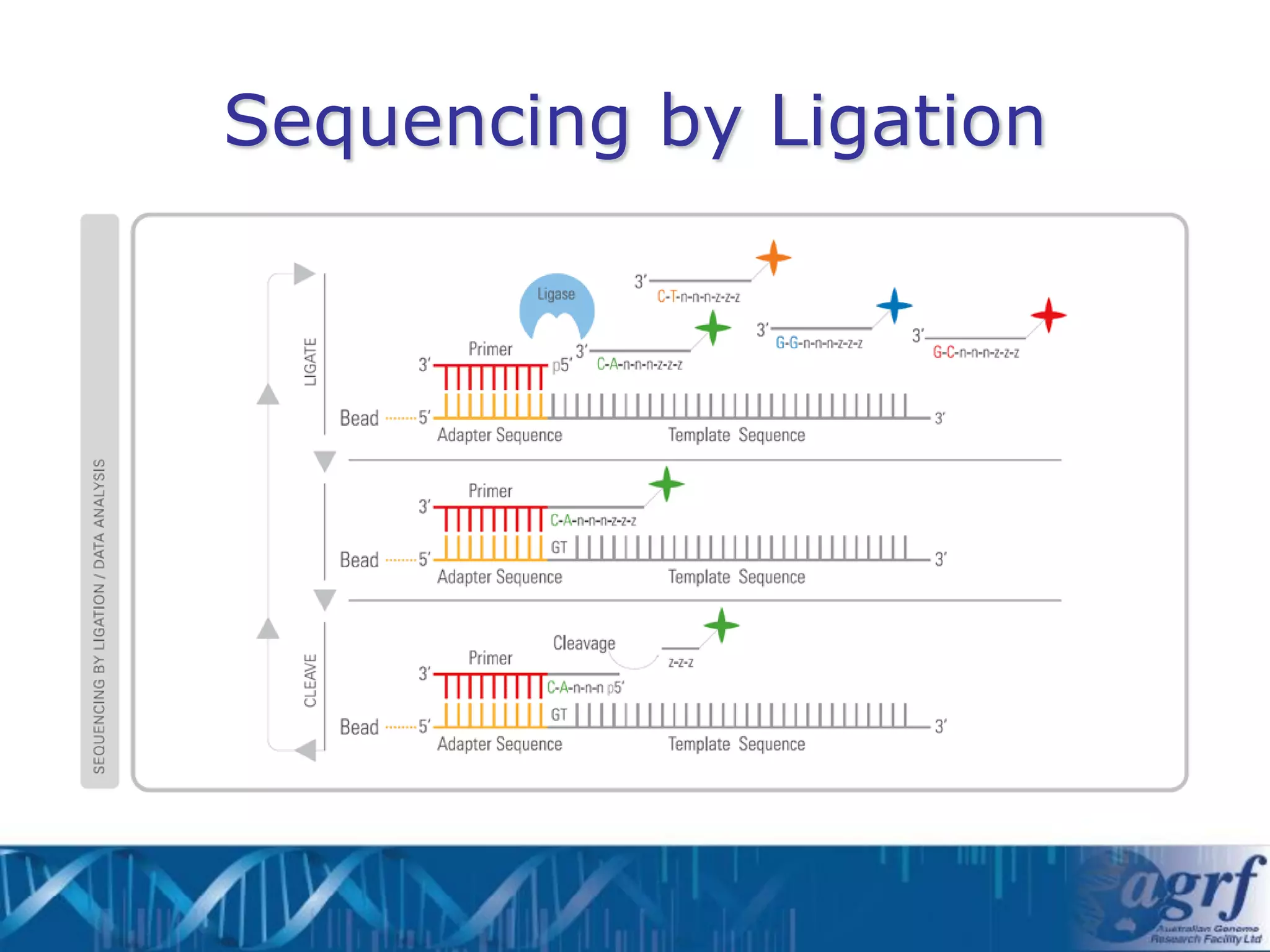
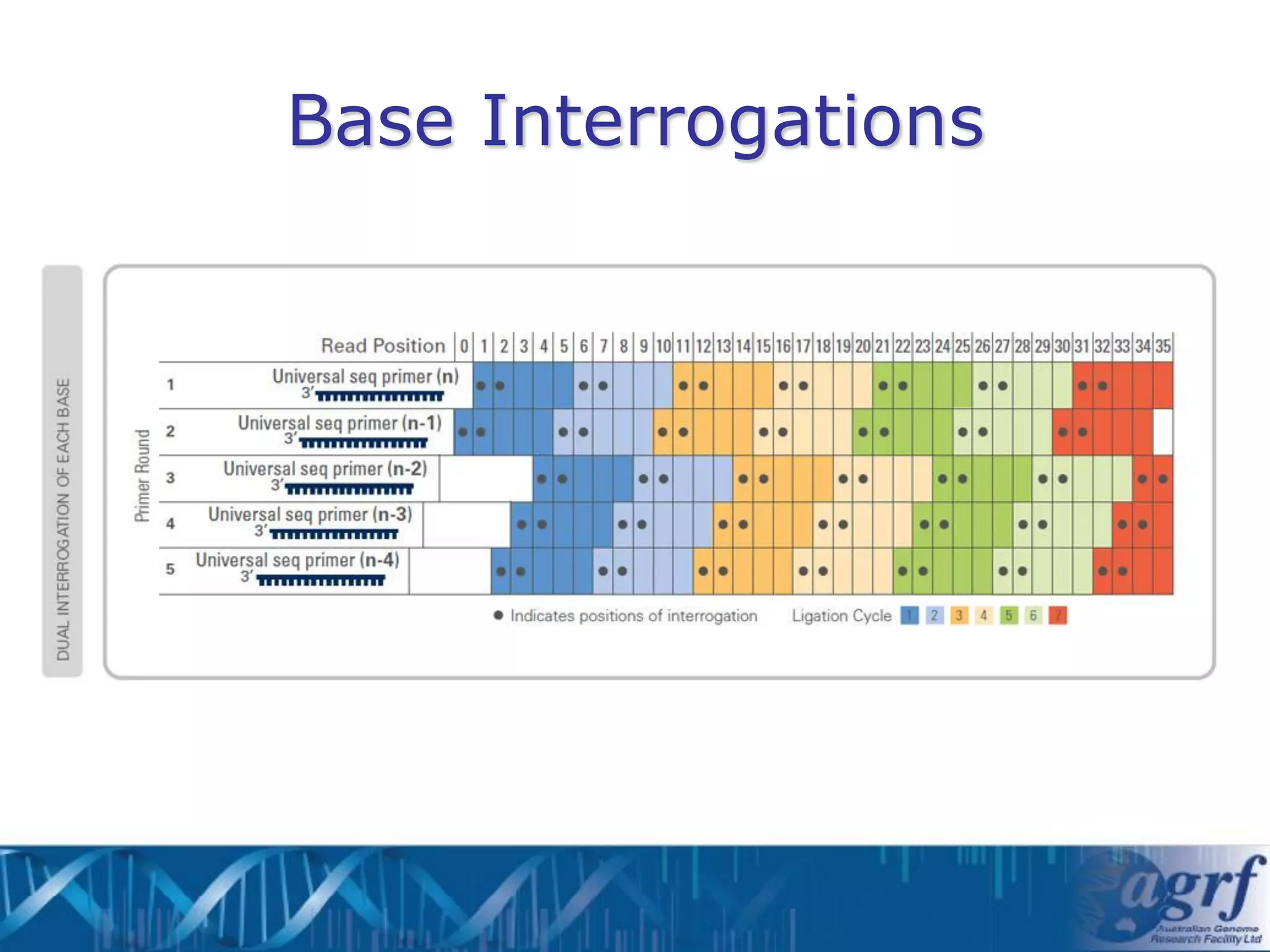
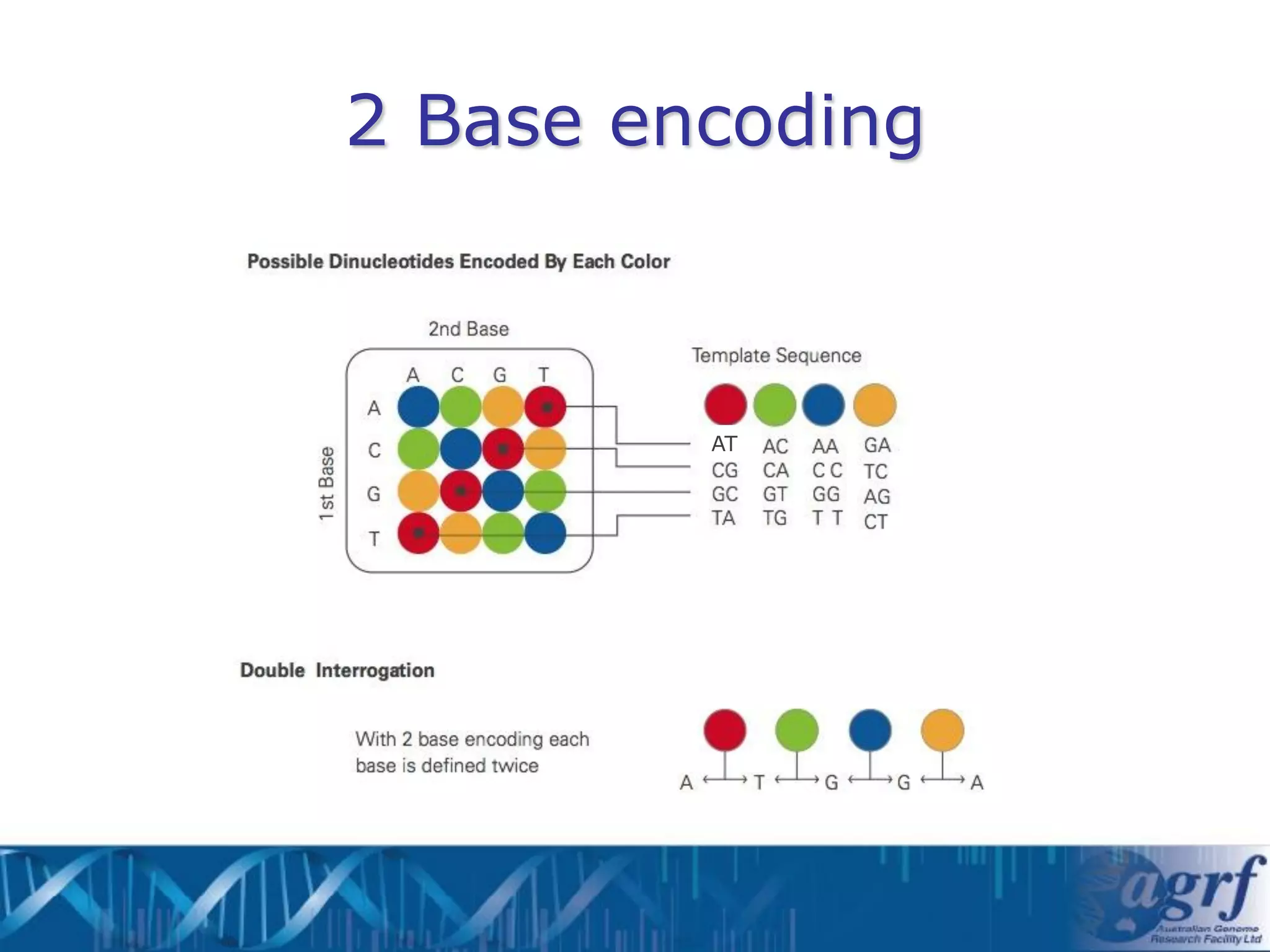
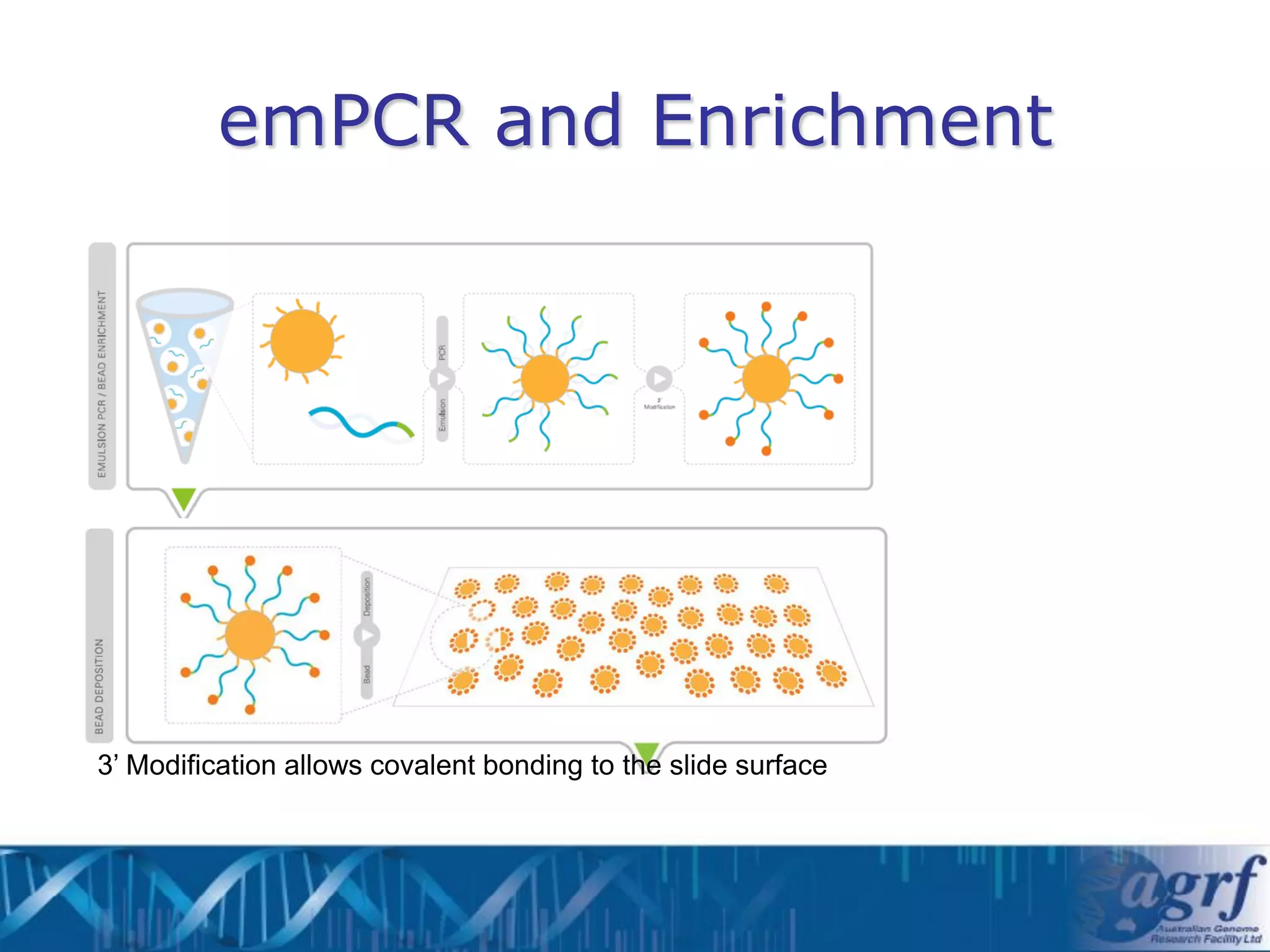
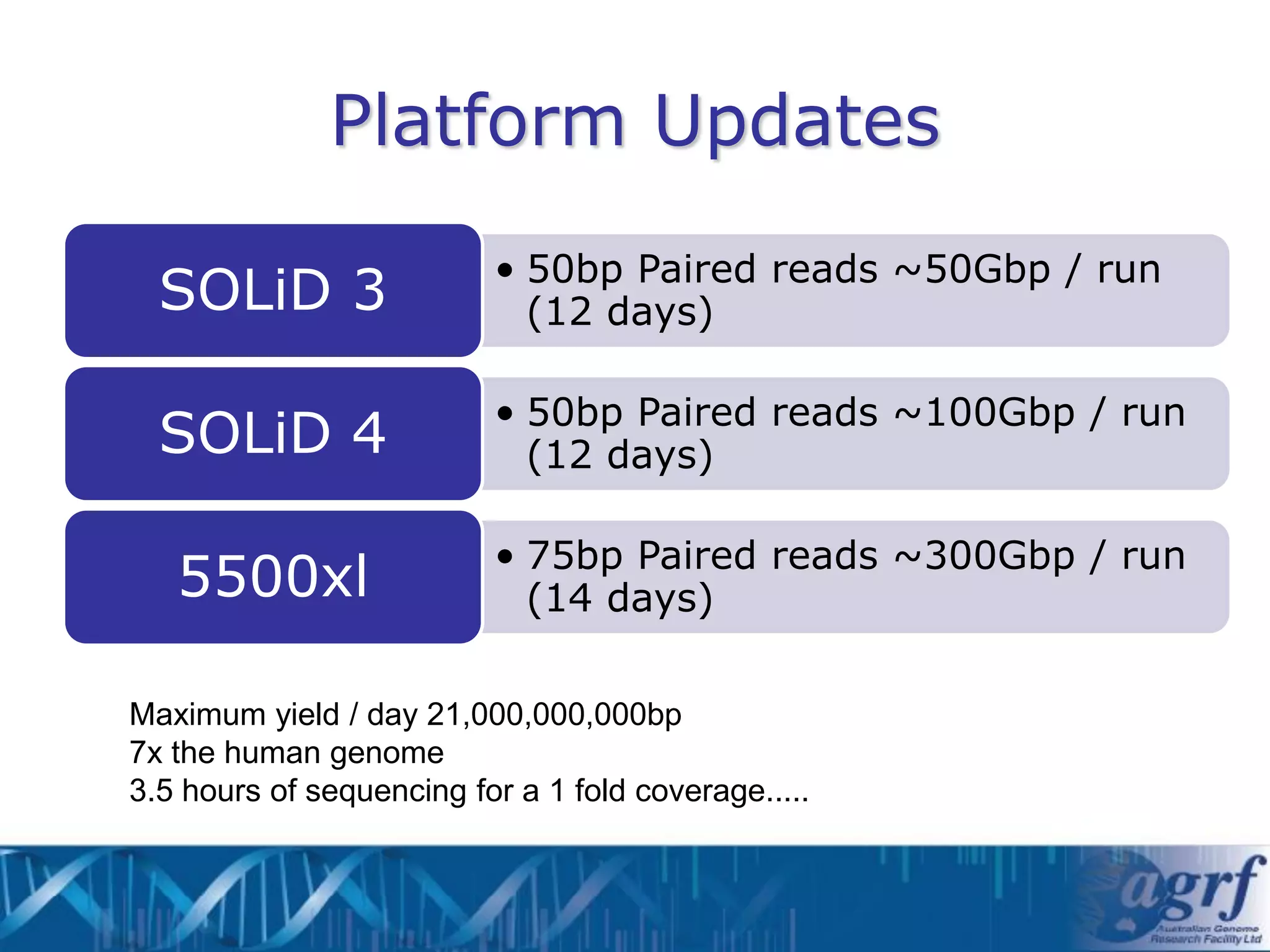
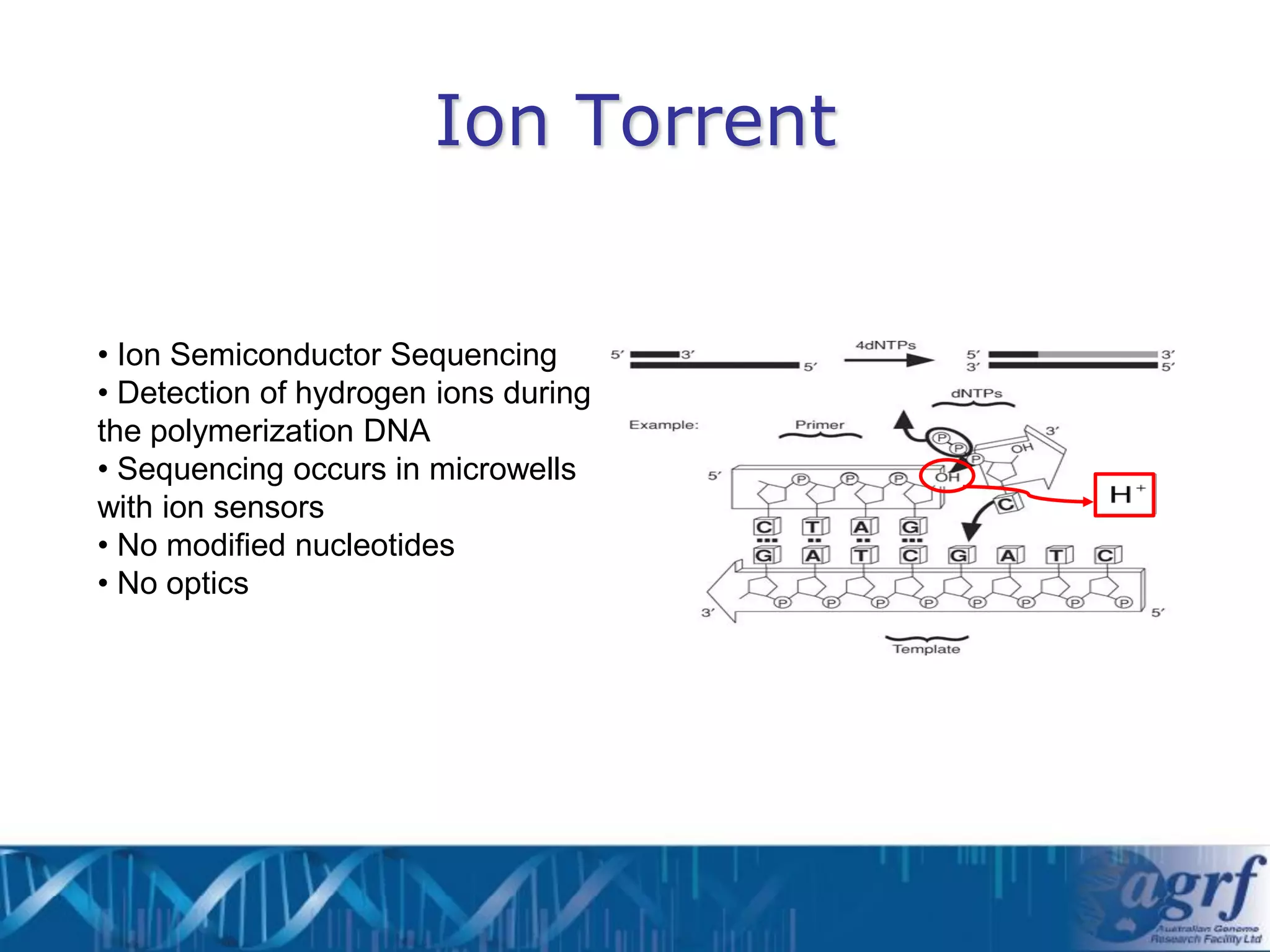
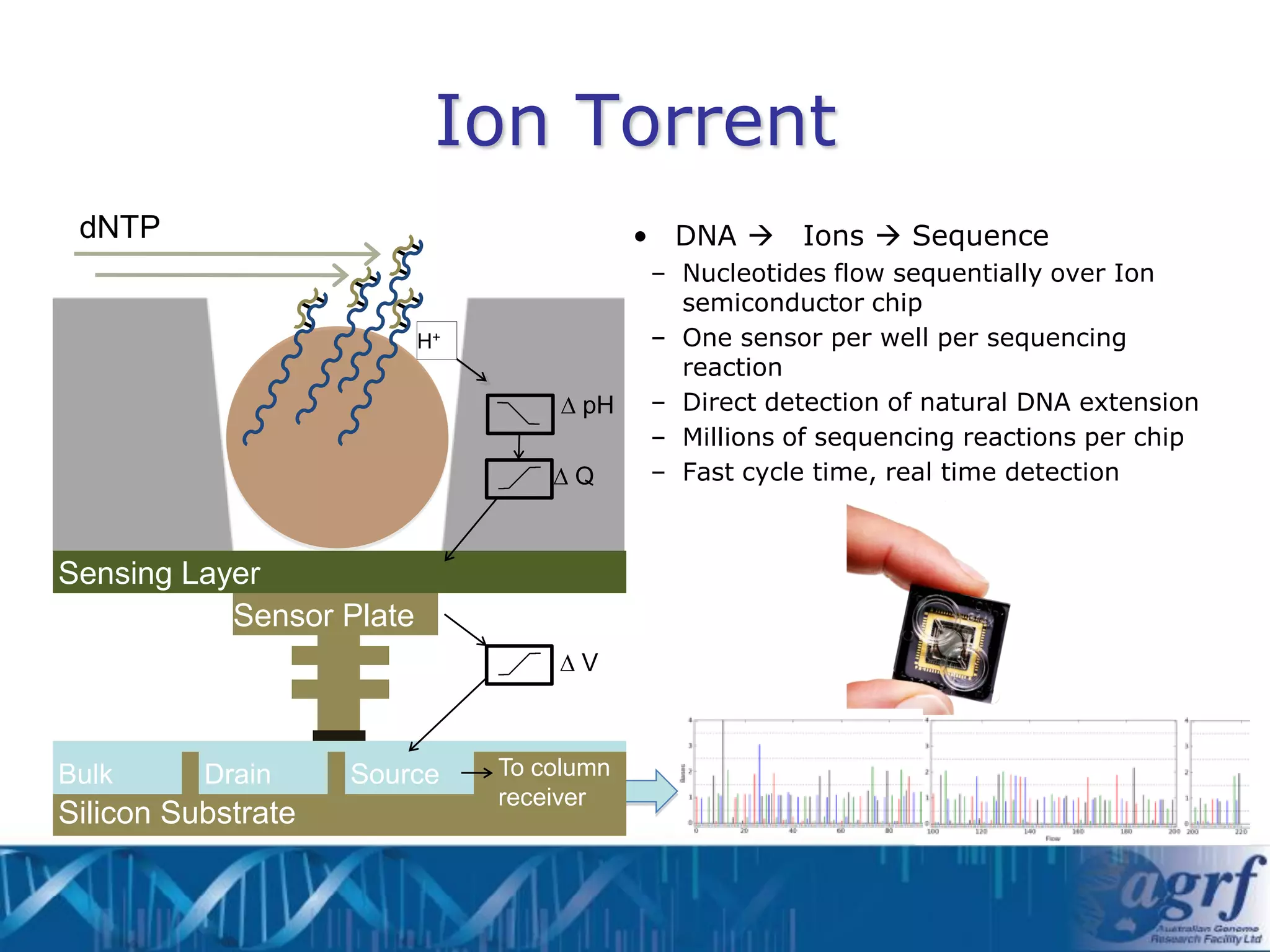
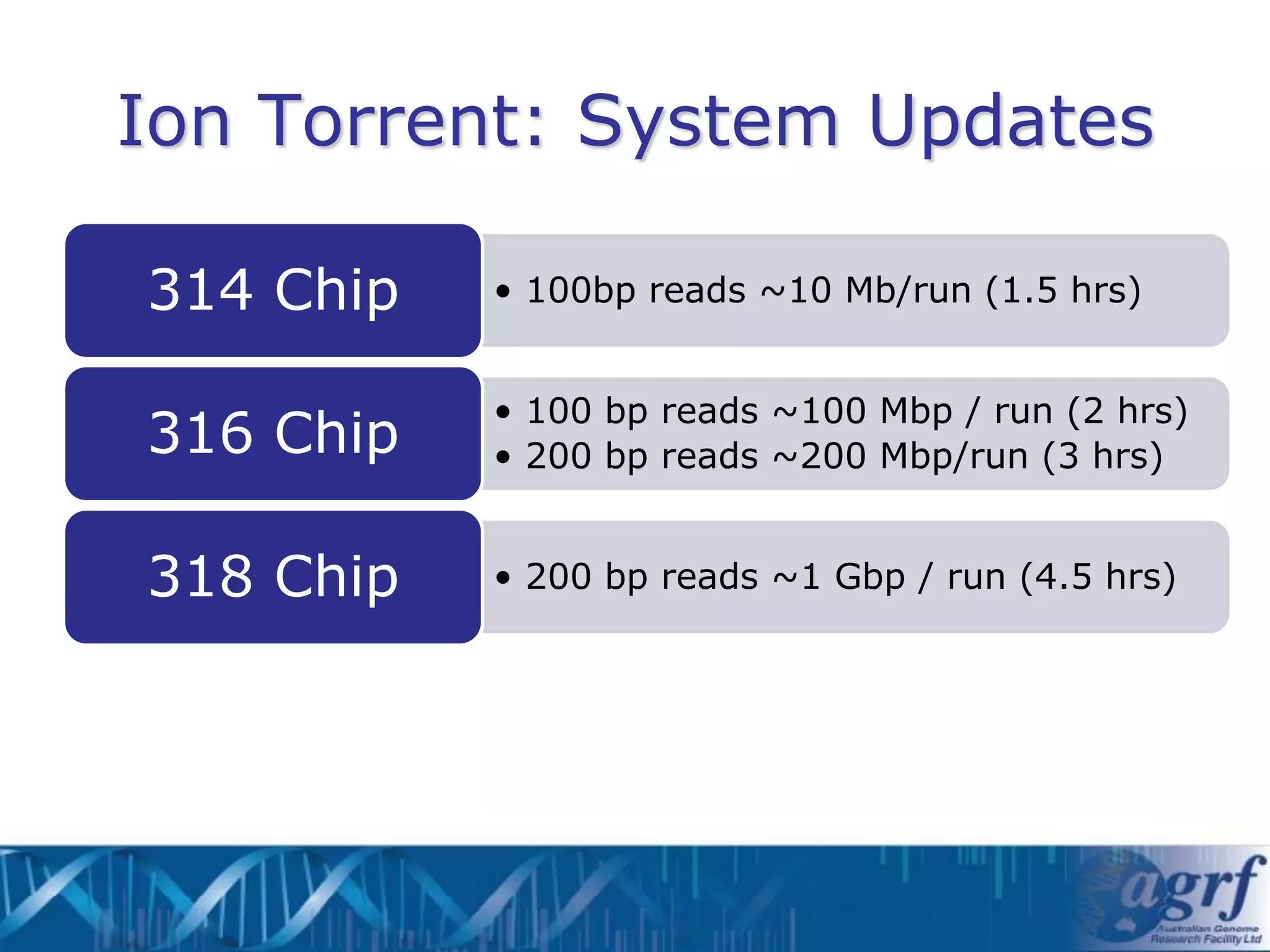
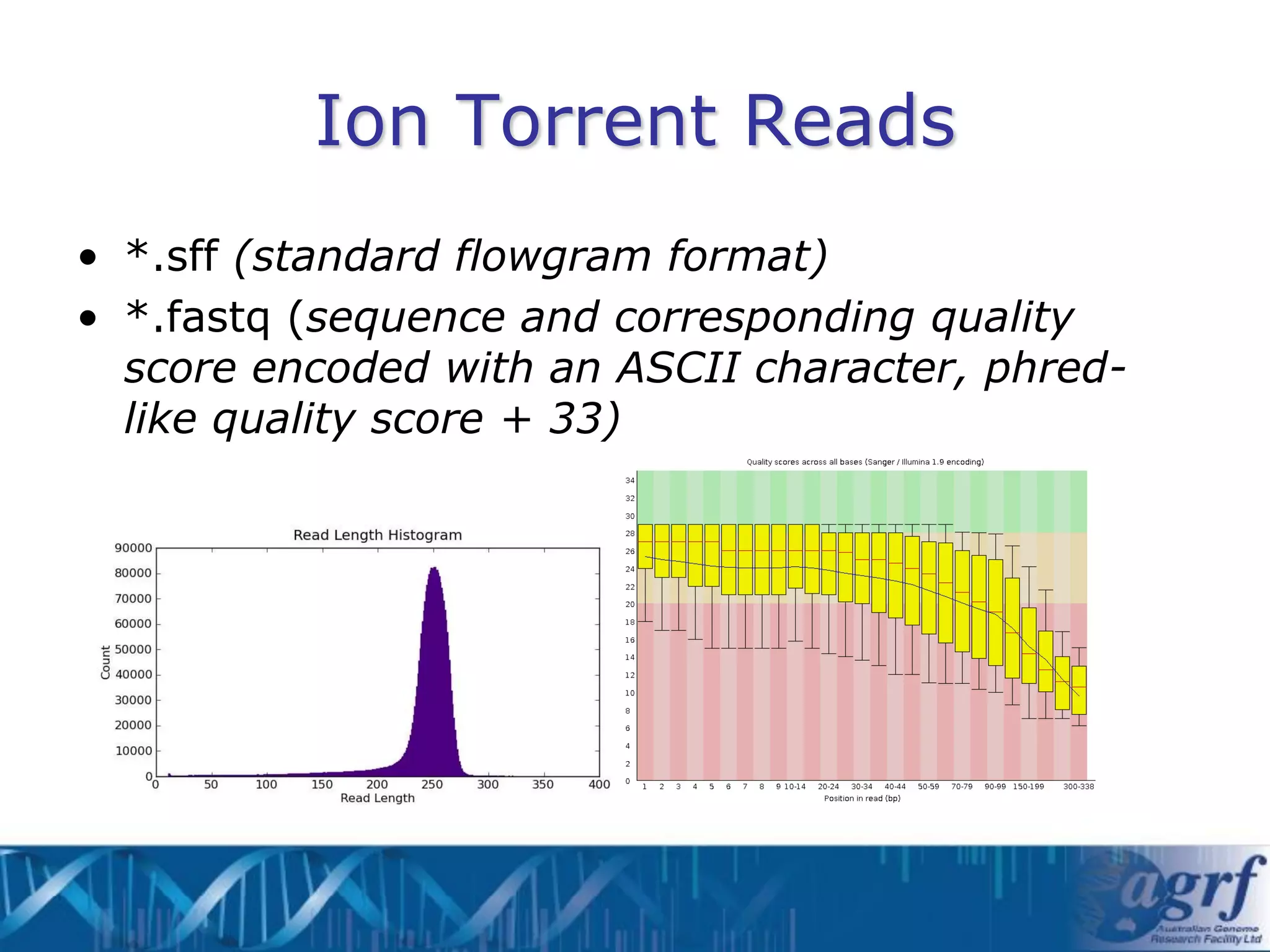
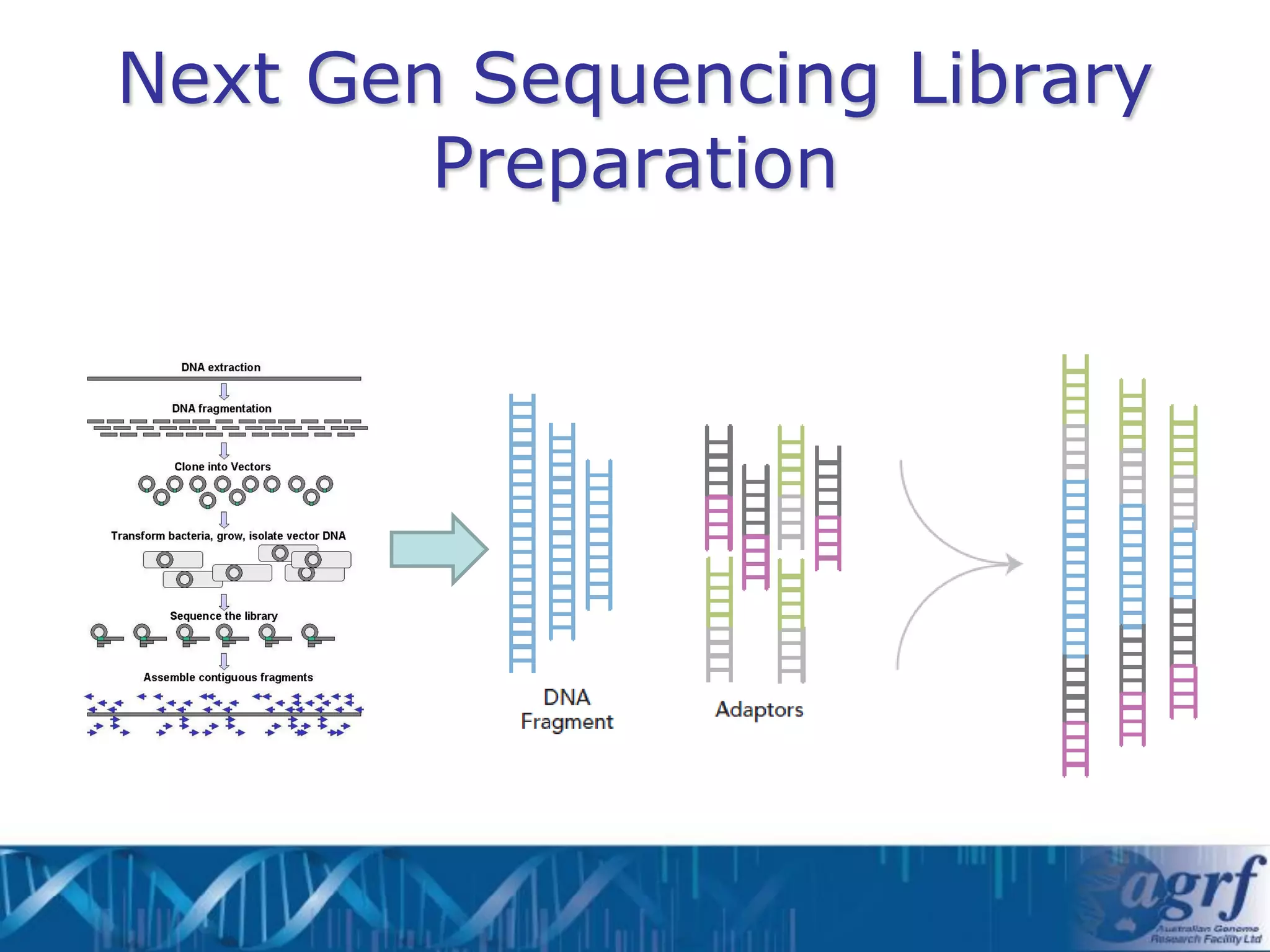
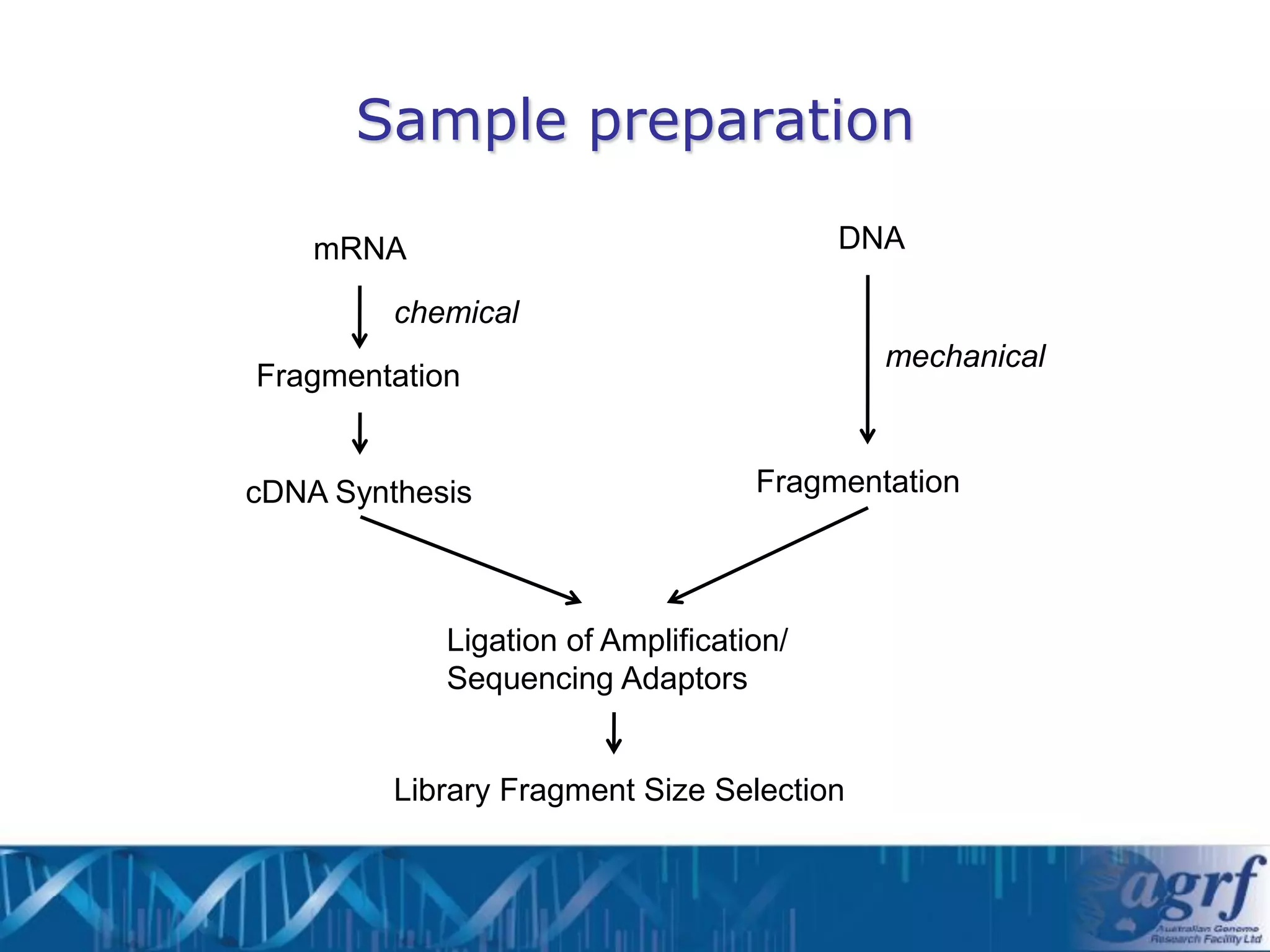
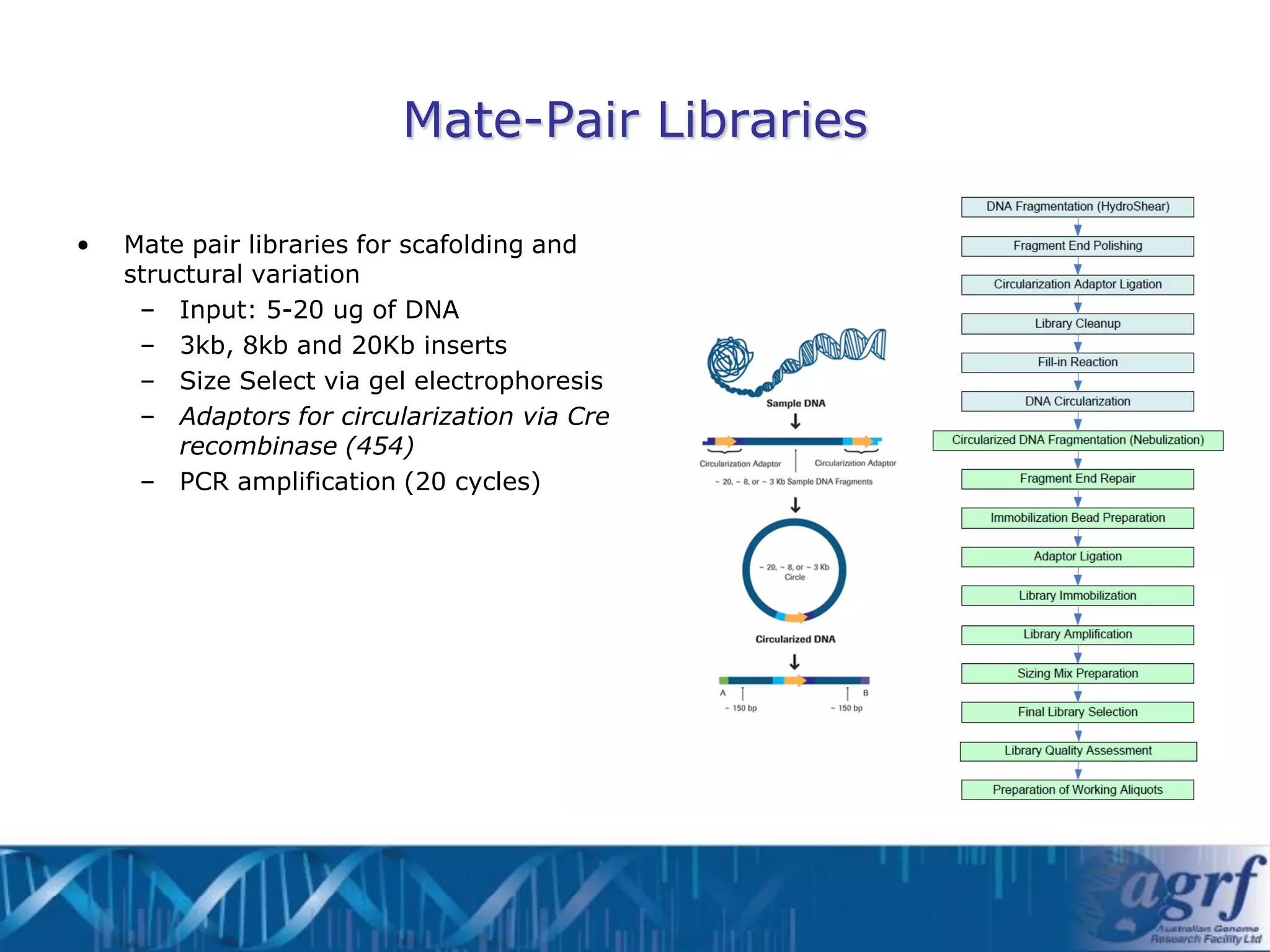
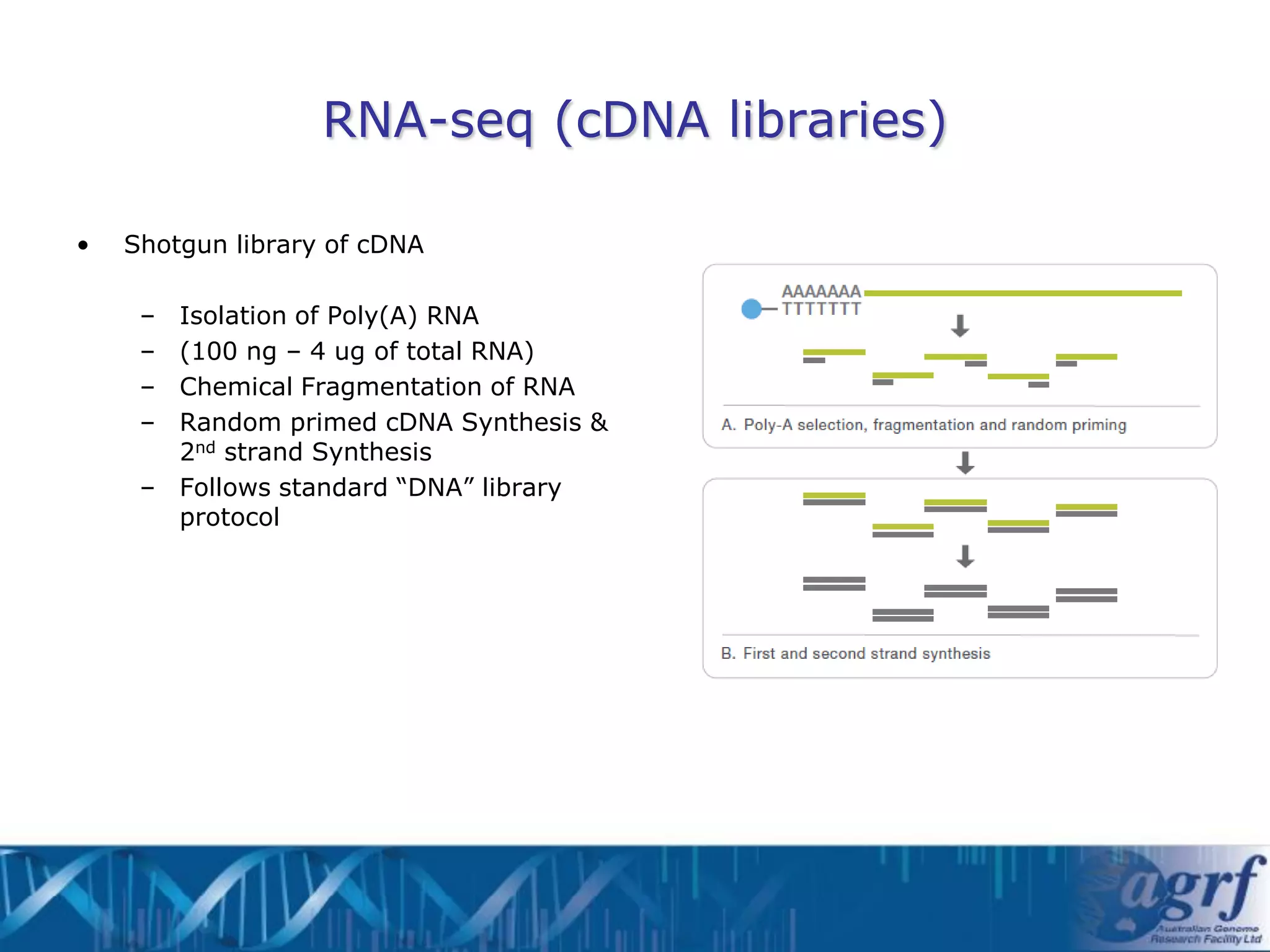
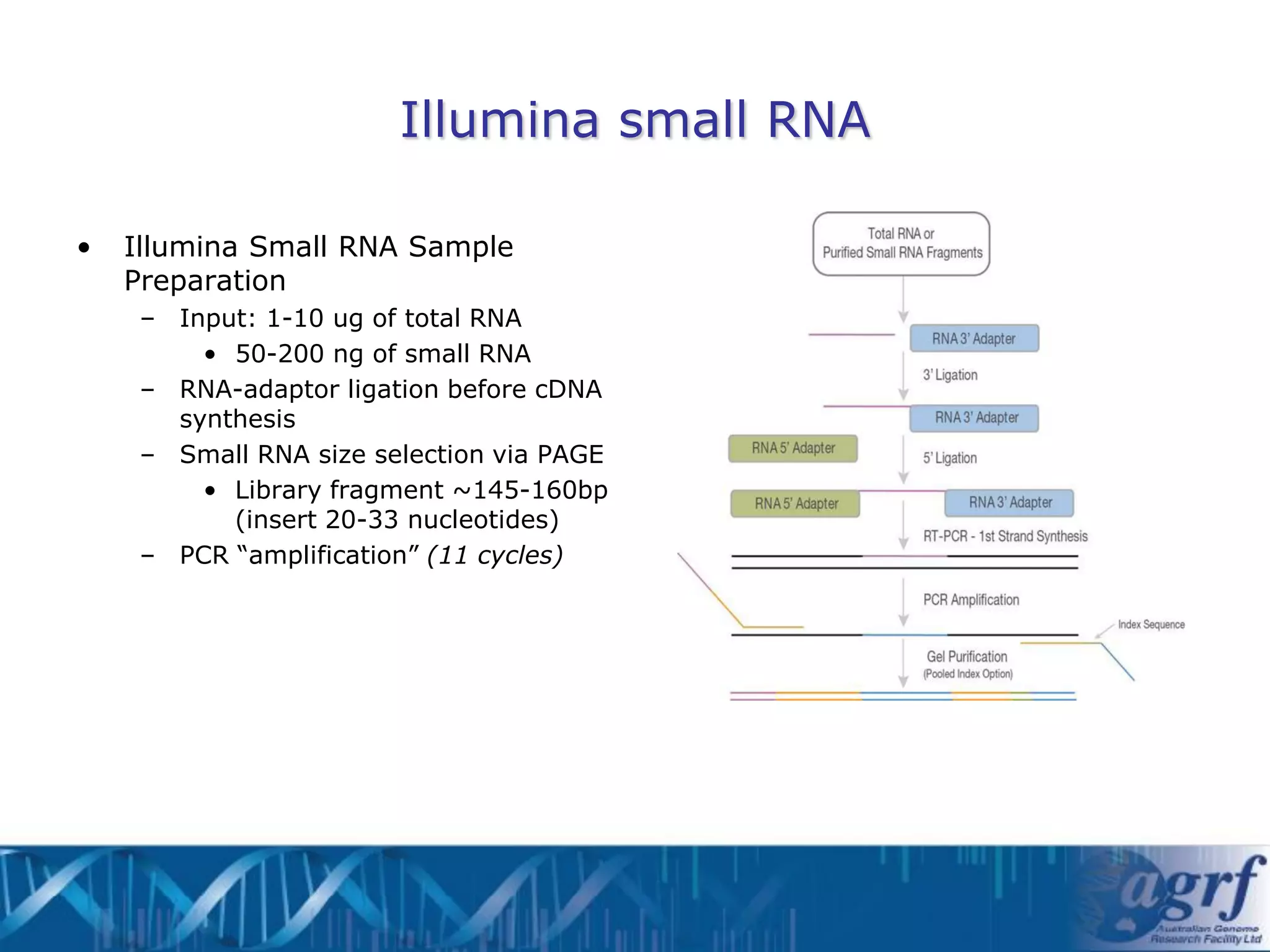
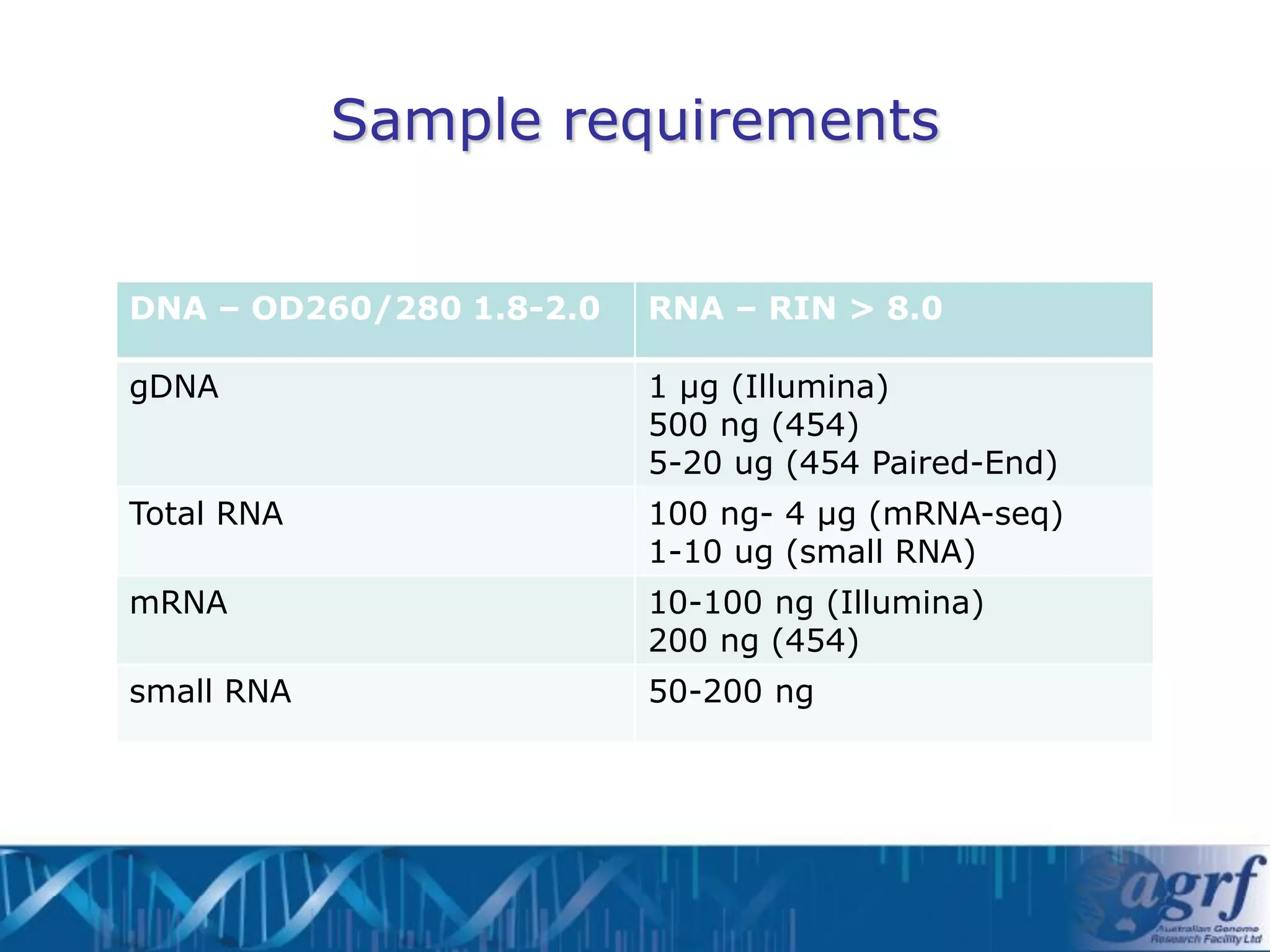
This document summarizes several next-generation sequencing platforms and applications. It describes the workflows and chemistries of 454, Illumina, SOLiD, and Ion Torrent platforms. These platforms have significantly reduced the cost of sequencing compared to Sanger sequencing. Common applications include whole genome sequencing, RNA sequencing, sequence capture, and amplicon sequencing. Library preparation requires fragmentation of DNA or RNA, addition of adapters, and amplification prior to sequencing.