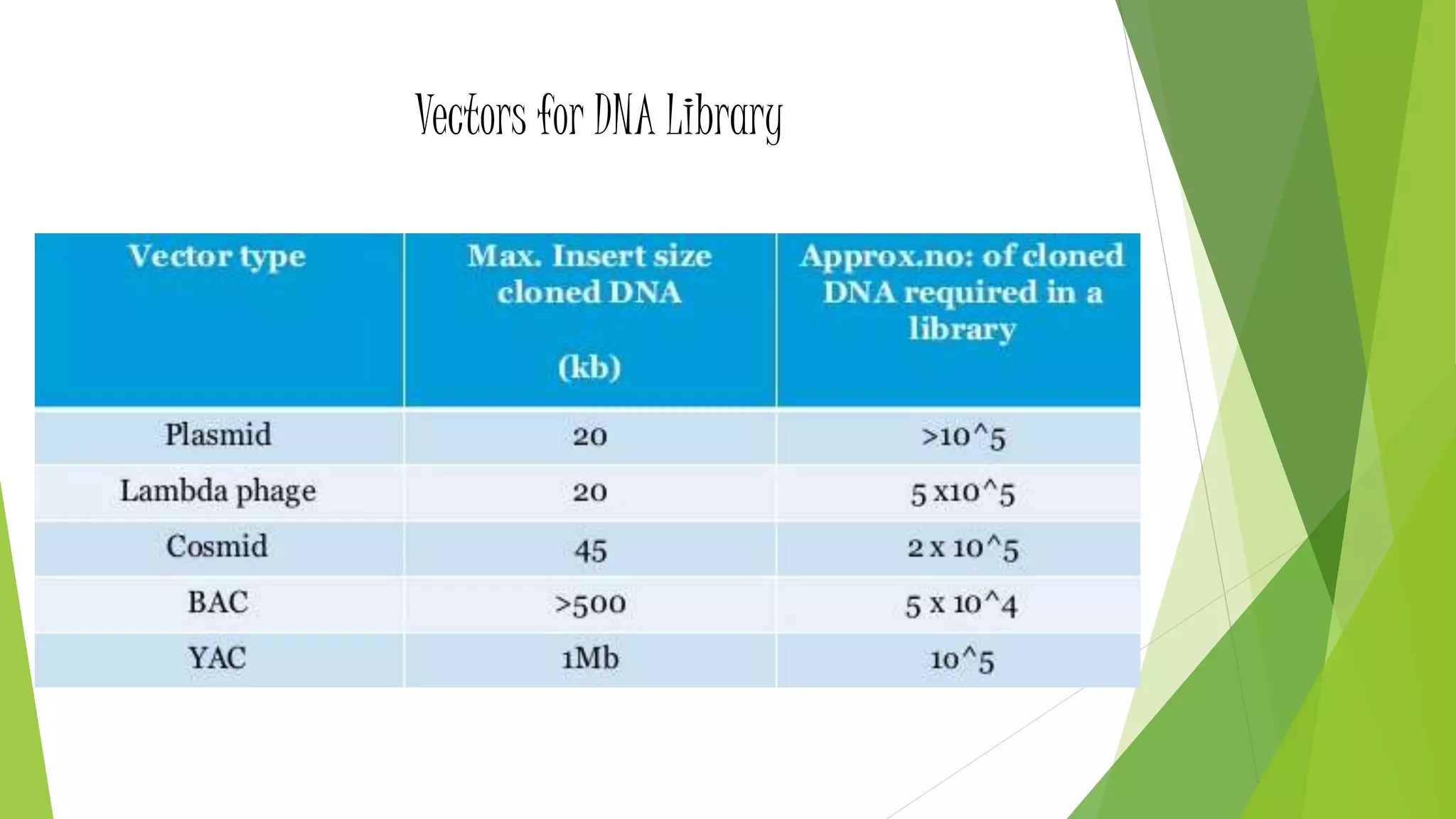
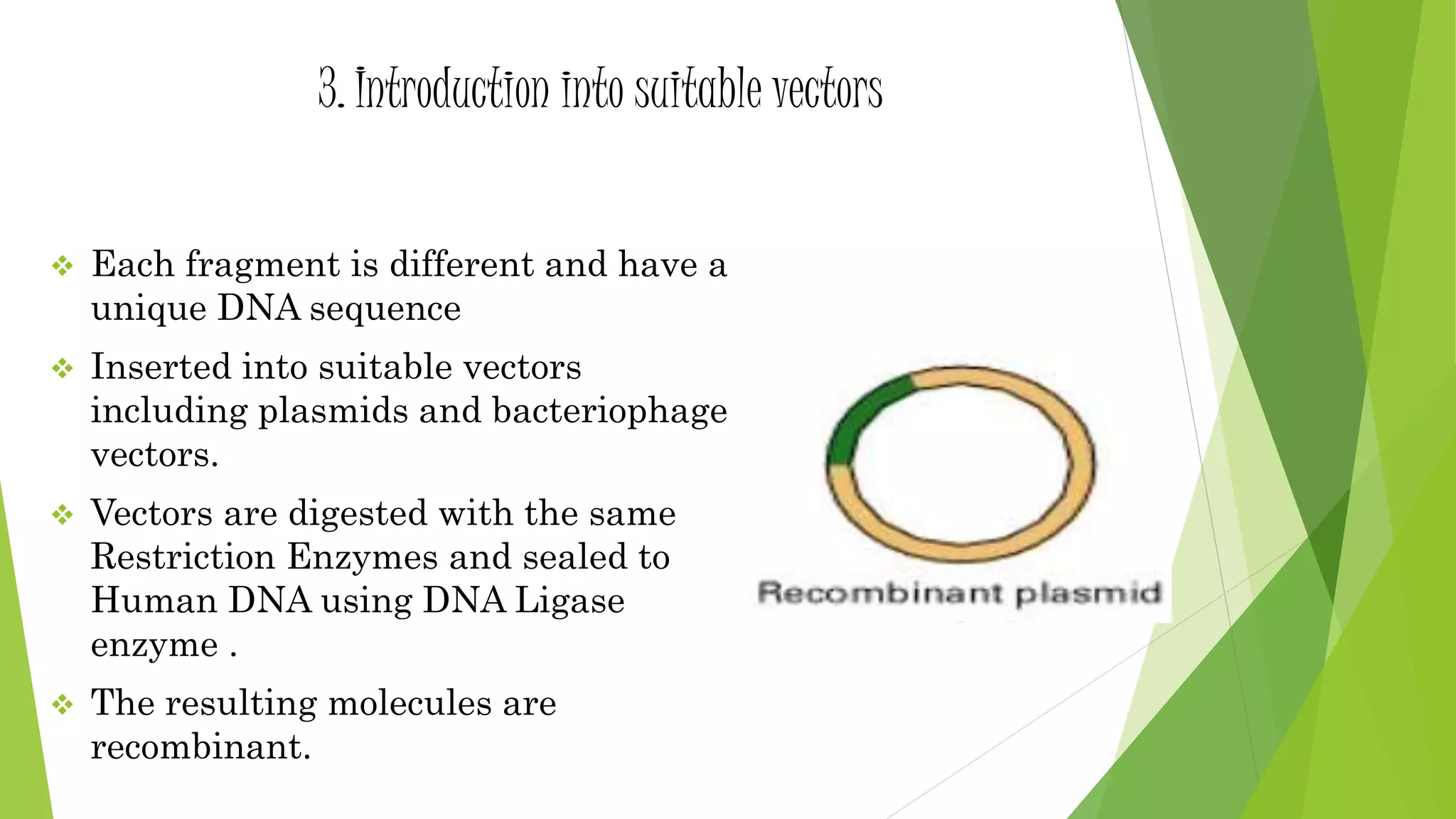
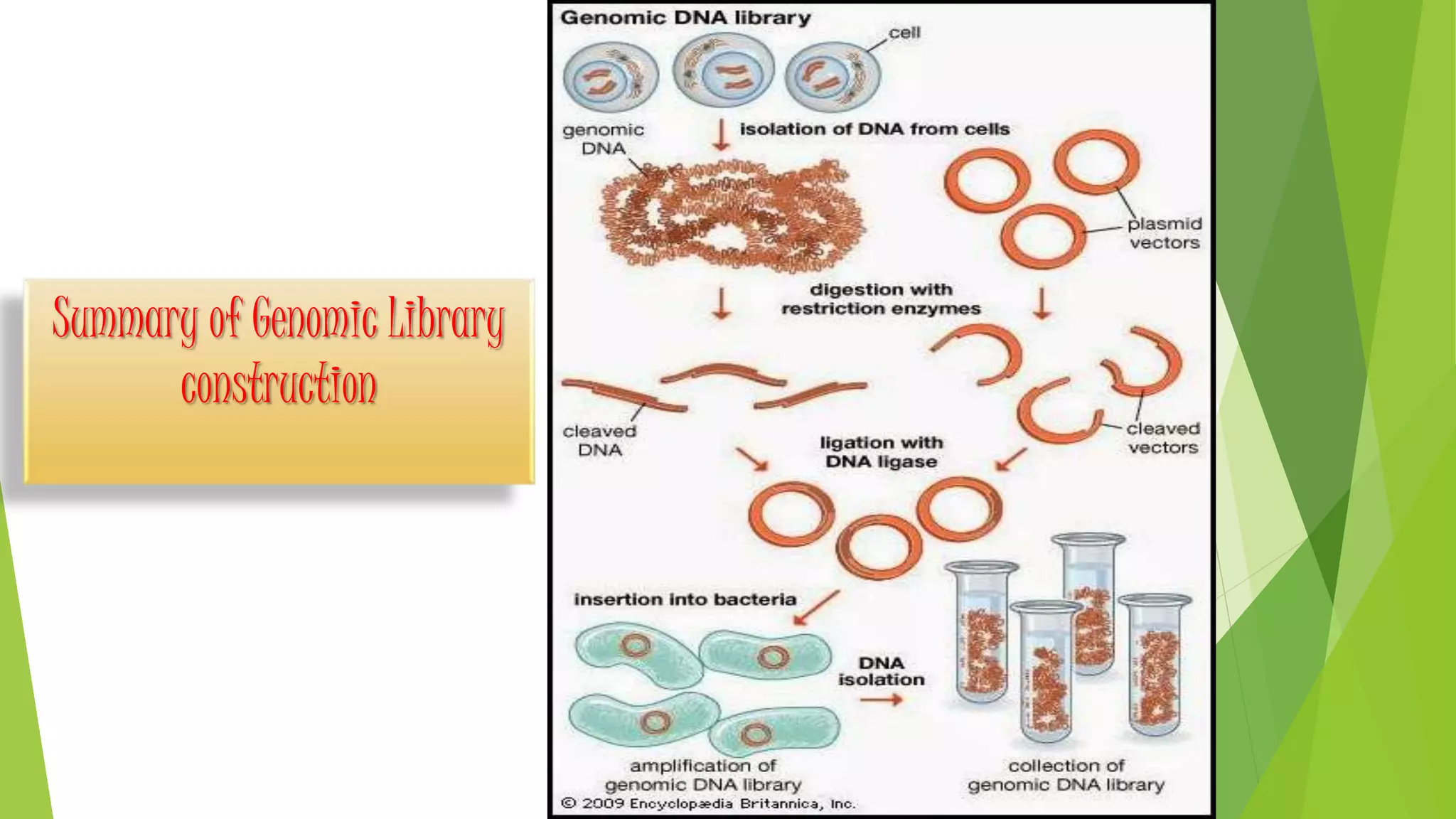
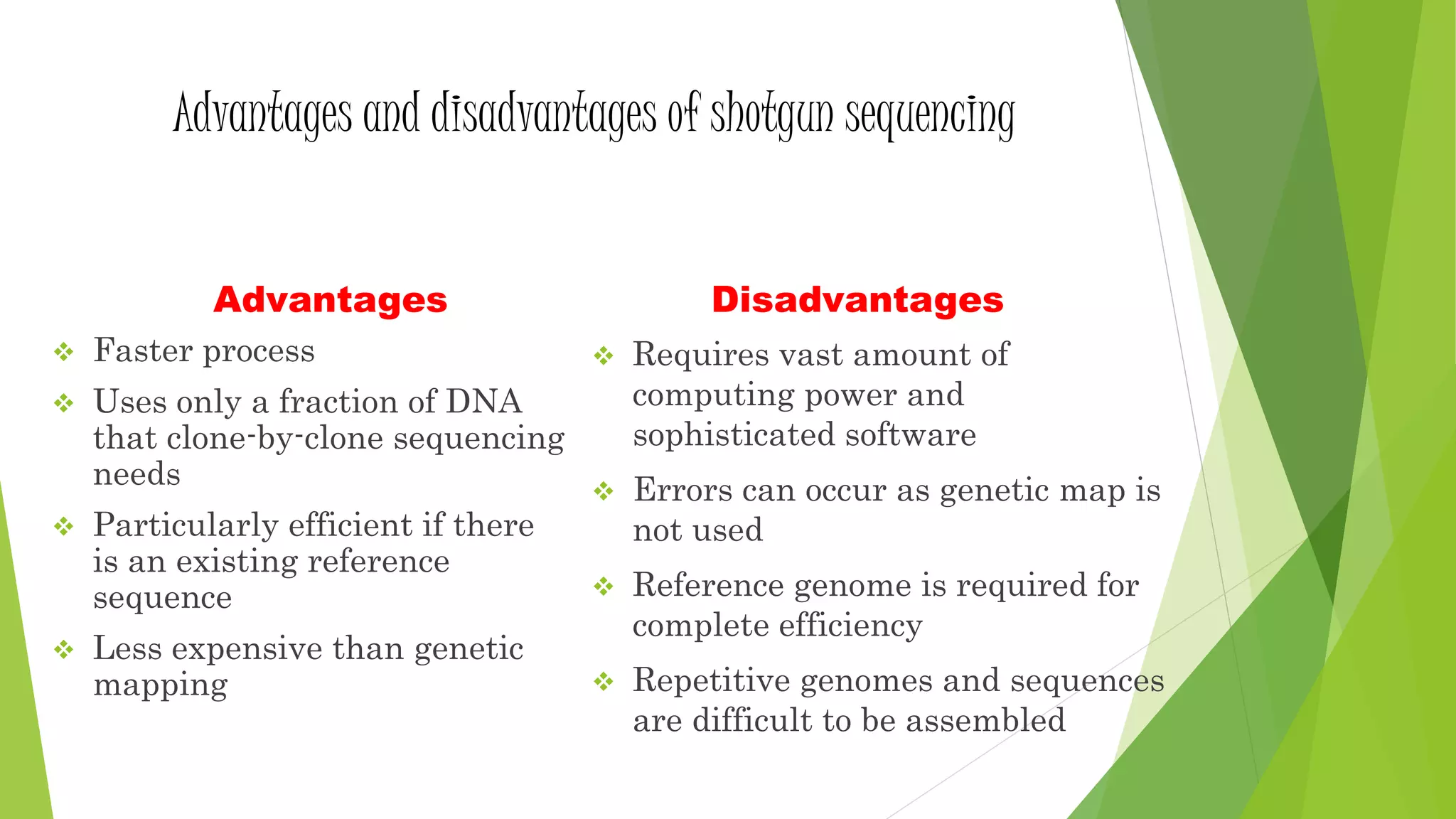
The document provides an overview of genomic libraries, detailing the methods of gene isolation and cloning, and discusses two types of DNA libraries: genomic and cDNA. It explains the construction process of a genomic library, including isolation, digestion, and insertion of DNA into vectors, as well as screening methods for selecting recombinants. Additionally, the text highlights the uses of genomic libraries in research, such as exploring genomic structure and function, and introduces shotgun sequencing techniques.