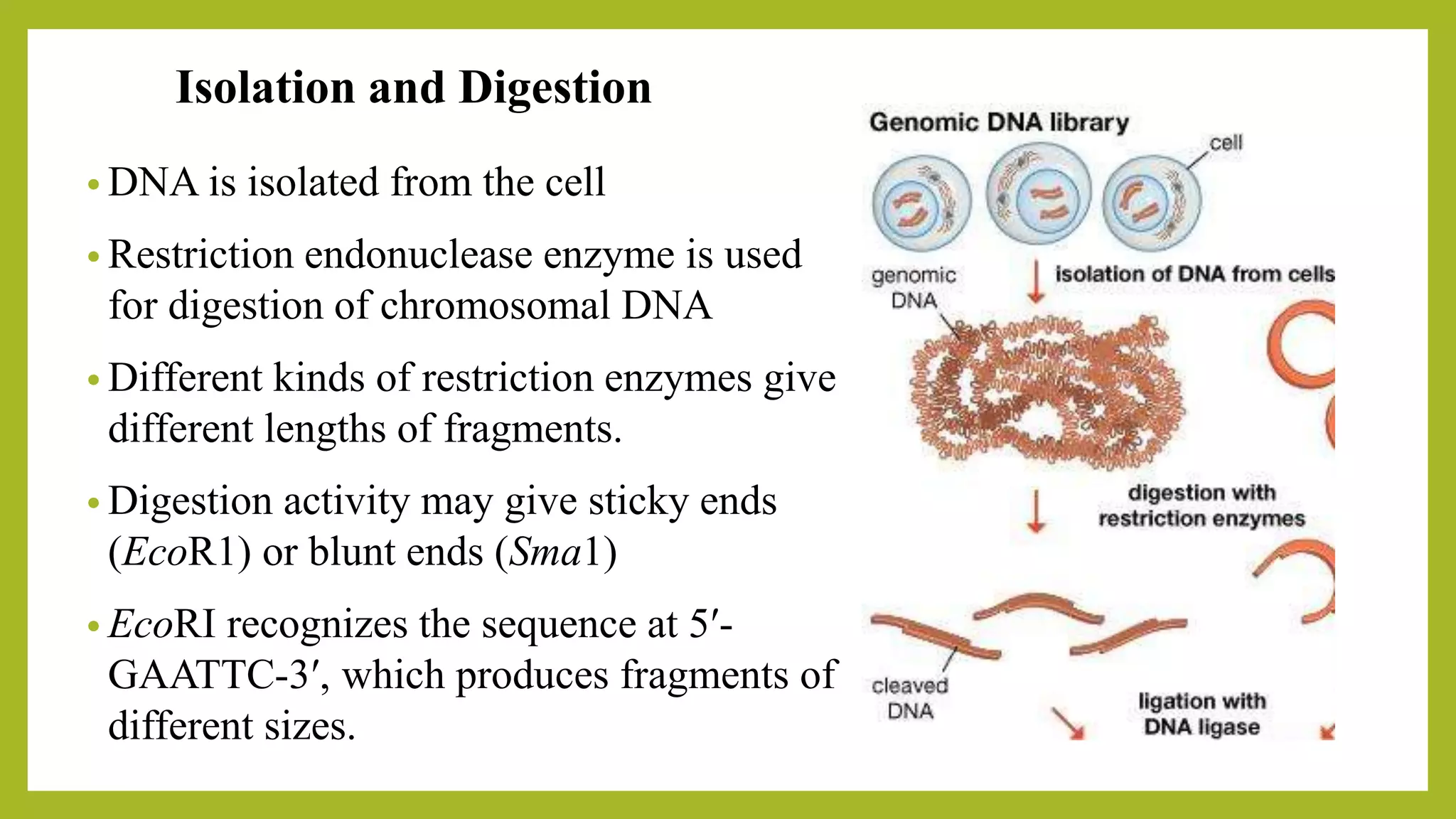
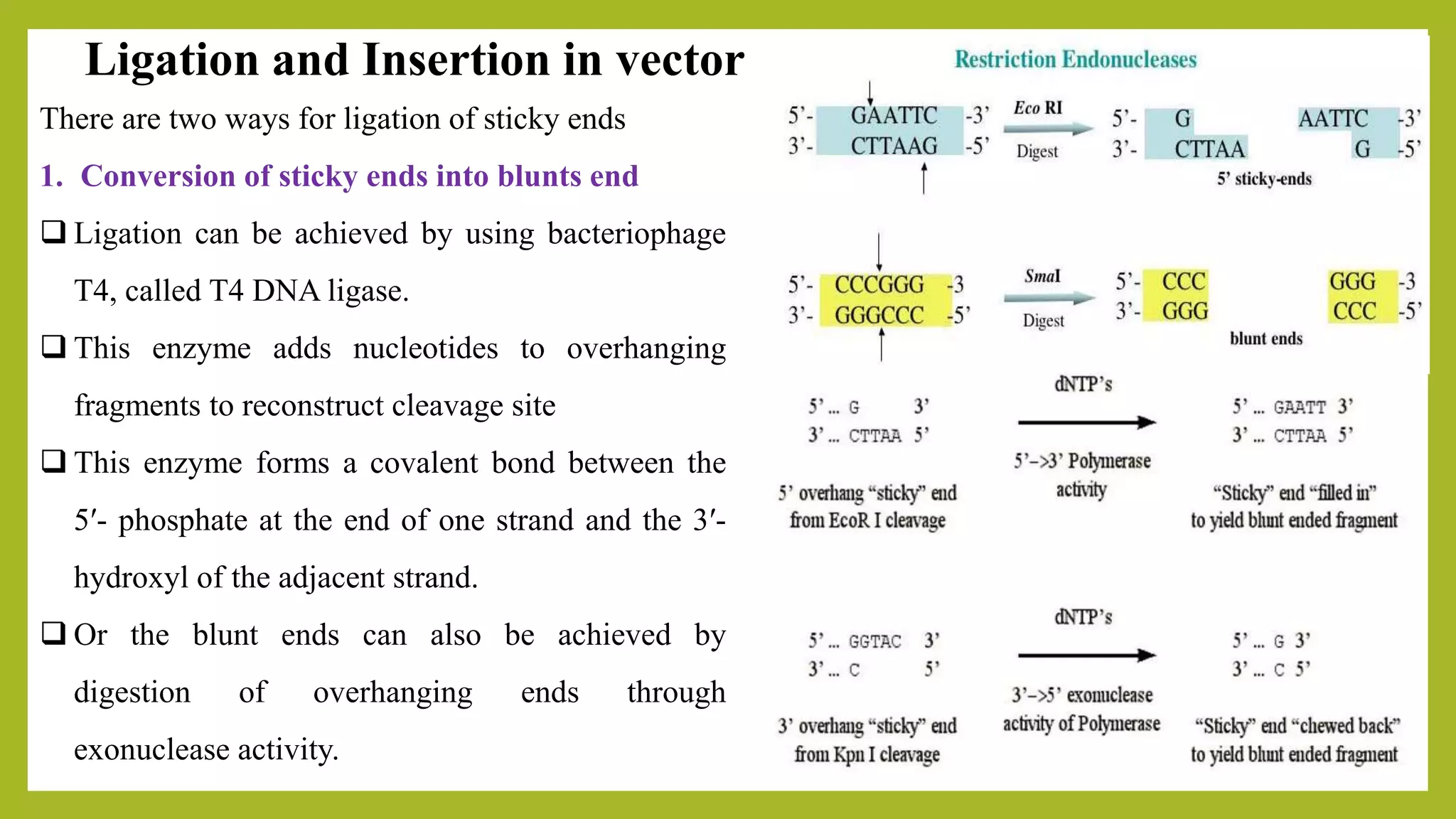
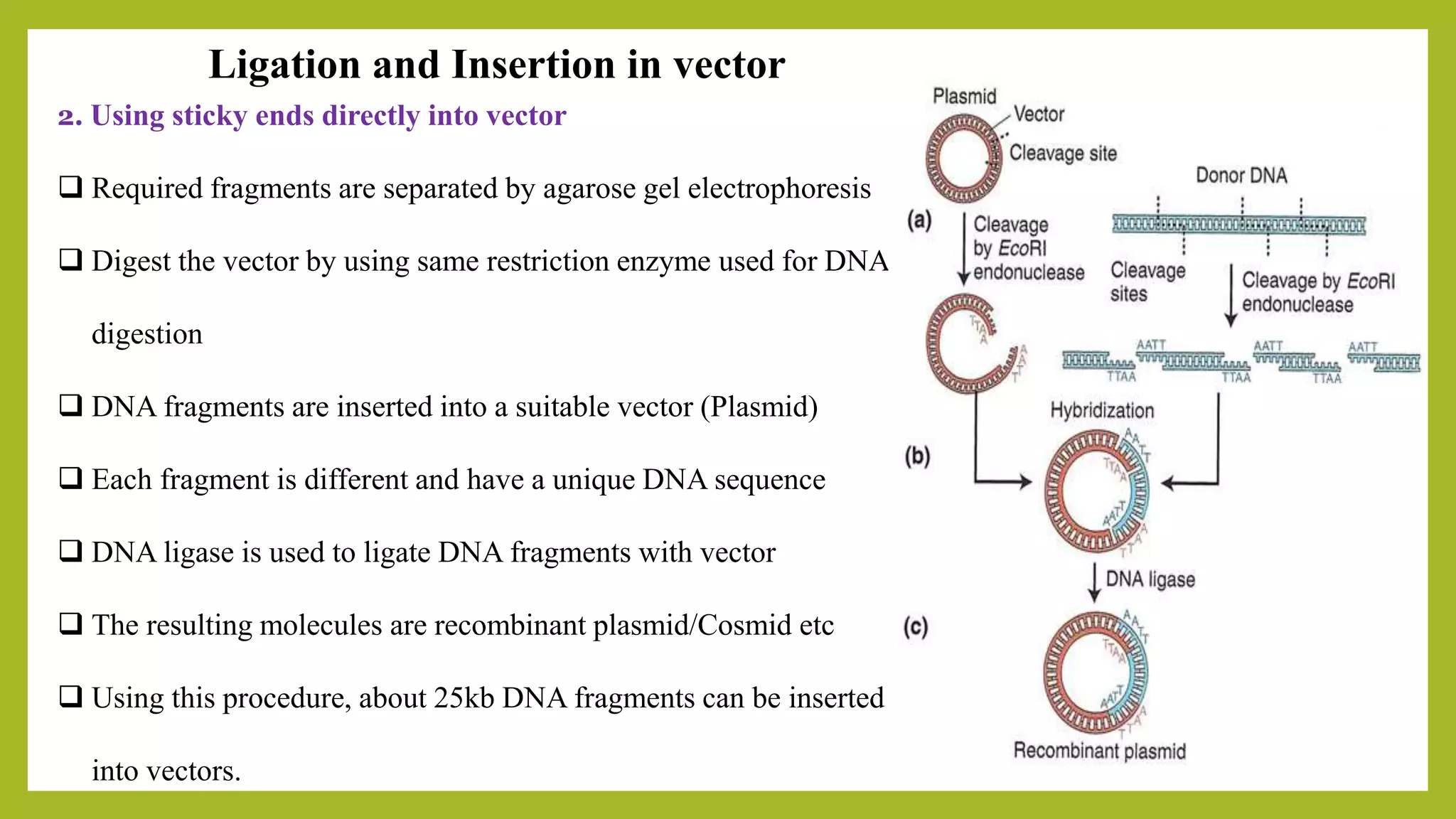
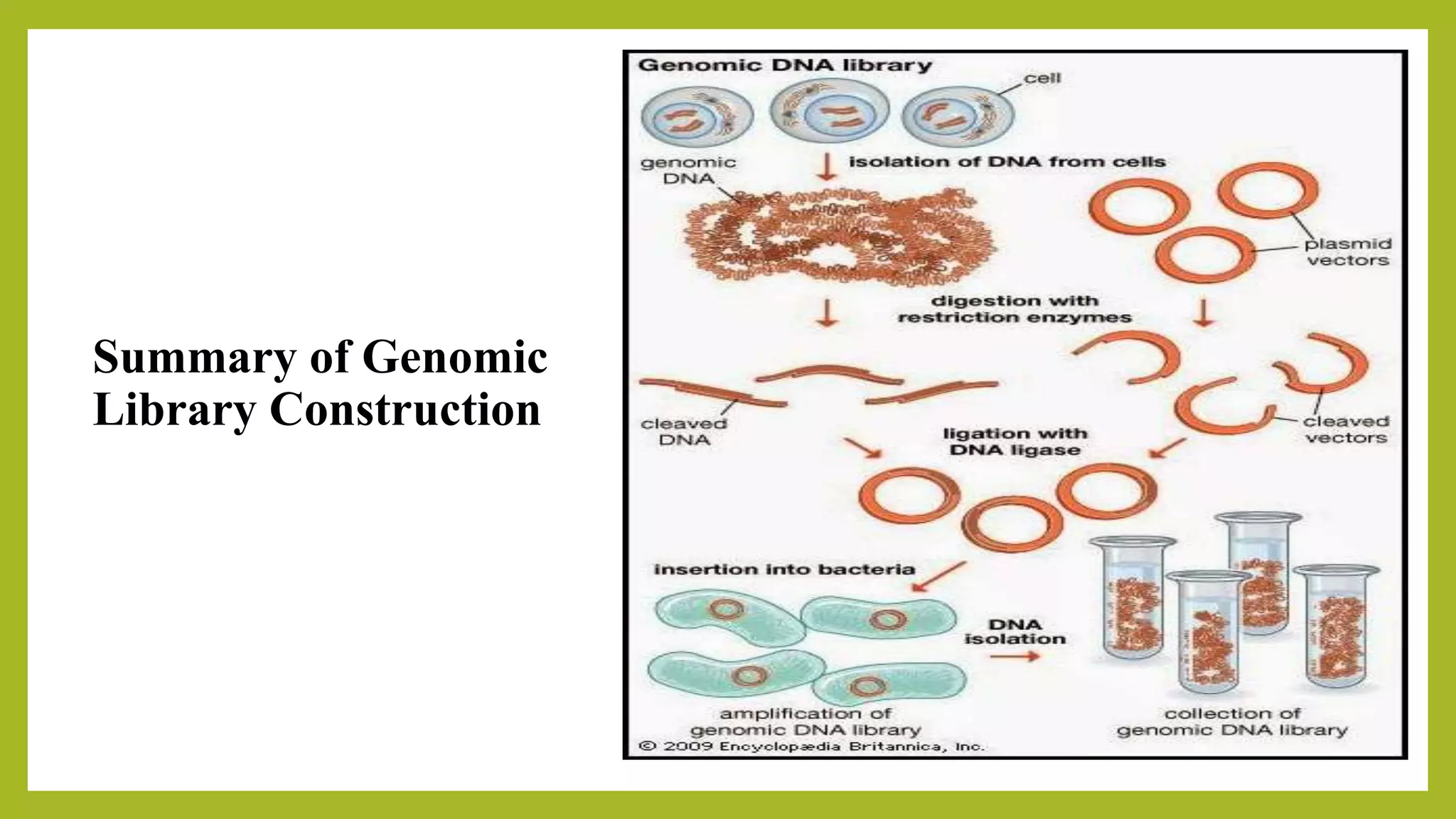
The document discusses genomic libraries, which are collections of DNA fragments representing an organism's entire genome, created through molecular cloning. It describes the importance of genomic libraries for exploring genetic structure, mapping genes, and studying mutations, as well as the steps involved in their construction and the challenges faced. Additionally, various screening strategies for gene libraries are highlighted.