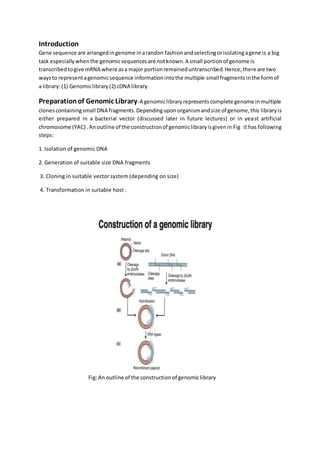
Genomic DNA library construction involves isolating genomic DNA from an organism and fragmenting it into smaller pieces to clone into a vector system. This creates a genomic library that represents the complete genome across multiple clones of small DNA fragments. Key steps include isolating high molecular weight genomic DNA, fragmenting it using restriction enzymes or shearing, cloning the fragments into a suitable vector like a bacteriophage or BAC depending on size, and transforming the clones into a host to generate colonies containing fragments of the genomic DNA. Genomic libraries allow researchers to explore and map genomes, identify genes, and enable applications like recombinant DNA technology and sequencing projects.