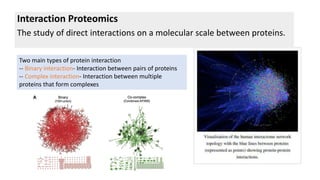
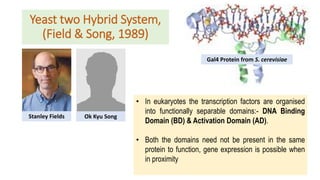
Protein interaction proteomics is the study of direct interactions between proteins on a molecular scale. There are two main types of protein interactions: binary interactions between pairs of proteins and complex interactions between multiple proteins that form complexes. Methods to study protein-protein interactions include genetic methods, bioinformatic methods, affinity-based biochemical methods, and physical methods like X-ray crystallography and NMR spectroscopy. The yeast two-hybrid system is an in vivo technique that detects interactions through reconstitution of a transcription factor when two proteins of interest interact.