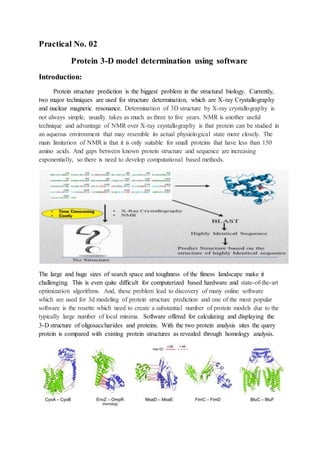
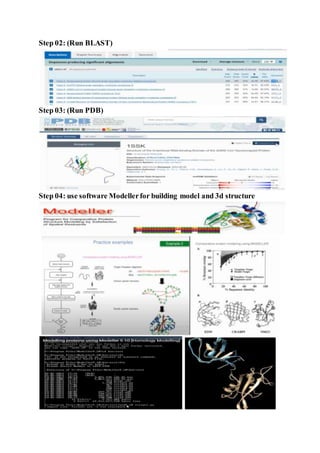
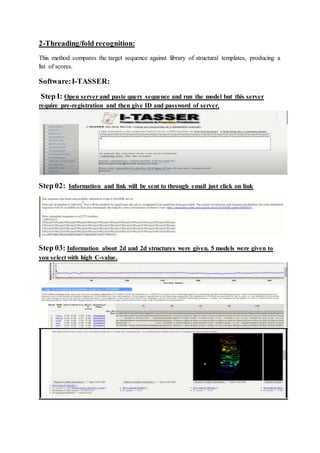
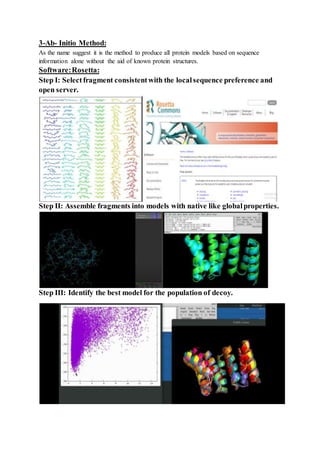
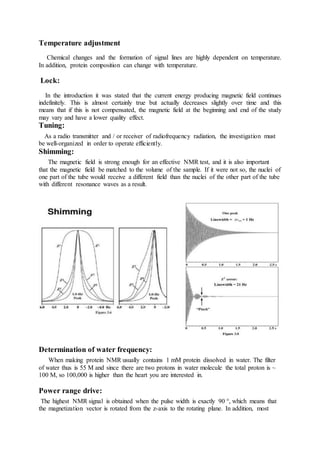
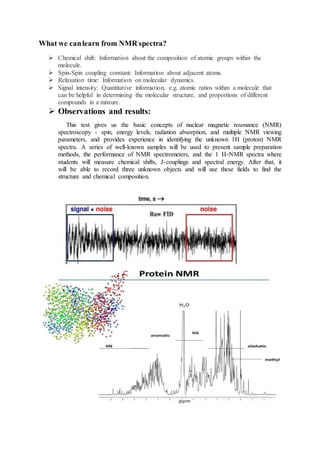
The document discusses protein 3D structure determination using computational modeling software. It describes different computational modeling methods like homology modeling, threading/fold recognition, and ab initio modeling. Homology modeling involves comparing the target sequence to known protein structures while threading/fold recognition compares the target to known structural templates. Ab initio modeling produces structures based only on the sequence. Popular software tools for each method are discussed like Modeller, SwissModel, I-TASSER, and Rosetta. The document also provides an overview of using nuclear magnetic resonance (NMR) spectroscopy to study protein structures experimentally.