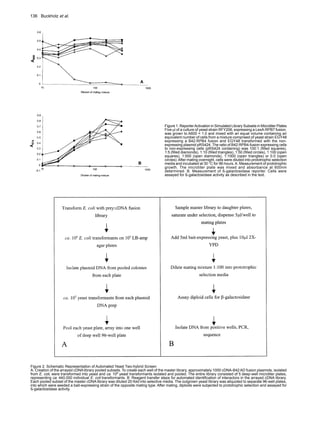
This document describes an automated method for yeast two-hybrid screening that pools cDNA library subsets in a liquid format. Key points:
- Yeast strains expressing "bait" proteins are mated with pooled subsets of a cDNA library expressed as "prey" fusions in 96-well plates. Interactors are selected and detected using reporters.
- Testing with simulated libraries containing varying ratios of interacting vs. non-interacting clones showed the method can detect interactions even when the interactor is only 0.1% of the prey pool.
- The method was used to screen two nuclear receptor ligand-binding domains against pooled subsets of two cDNA libraries, recovering both previously known and new interacting proteins.



![Yeast Two-Hybrid Screening 139
tandem mass spectroscopy (McCormack et al., 1997) is
facilitating the characterization and identification of
individual components of multisubunit complexes at the
biophysical level. In addition, DNA chip technology is
enabling the rapid identification of gene sequences
encoding interacting clones isolated in Y2H screens (Cho
et al., 1998). We anticipate that the coordinated use of
these techniques, with automated yeast two-hybrid
screening and differential gene expression, will make it
possible to build a database of genes that are both co-
regulated and in the same biochemical and biophysical
pathways.
Experimental Procedures
Restriction and DNA modification enzymes were purchased from various
manufacturers and used according to their recommendations.
Creation of Arrayed cDNA Libraries
A human fetal brain yeast two-hybrid cDNA library (Invitrogen) was
transformed into E. coli and plated at a density of approximately 1000 clones
per plate onto 440 LB + Amp plates and incubated 1-2 days at 37 °C. Next,
3-4 ml of LB (containing 15% glycerol) was added to each plate and the
plates rocked on a platform shaker at low speed until suspension of the
colonies in the LB was apparent. A 200 µl portion of the suspended cells
was removed for plasmid DNA isolation.
Plasmid DNA was prepared by adding 250 µl of P2 solution (Qiagen)
to the 200 µl cell suspension in a 2 ml Eppendorf centrifuge tube. The two
solutions were mixed gently and then 250 µl P3 solution (Qiagen) was
added and the tubes shaken. The mixture was then centrifuged at 14K rpm
for 5 minutes in an Eppendorf centrifuge. The clarified supernatant (500 µl)
was pipetted to a fresh Eppendorf centrifuge tube and 1 ml of ethanol added
to precipitate the DNA. The precipitated DNA was centrifuged at 14K rpm
for 15 minutes, the ethanol solution decanted and the pellet dried in vacuo.
The pellet was dissolved in 50 µl sterile distilled water and used directly to
transform yeast.
Yeast were transformed using the EZ Yeast Transformation kit (Zymo
Research) according to the manufacturer’s recommendation, using 2.5 µl
DNA. The transformed yeast were incubated for 1 hr at 30 °C and plated
onto SC-W agar plates. The plates were incubated for an additional 3-4
days at 30 °C, and the cells harvested as for E. coli using 3-4 ml SC-W
containing 15% glycerol. The harvested yeast from each plate were
separately aliquoted into different wells of deep-well 96-well plates and
frozen in 15% glycerol at -80 °C, to create a master library.
Yeast Liquid-mating
Five µl from each yeast master library well were inoculated into 100 µl of
SC-W in individual wells of a 96-well plate and grown overnight at 30 °C.
Five µl of these overnight cultures were each transferred to a fresh
96-well V-bottom plate. A 5 µl aliquot of bait yeast culture (OD600 = 1) was
added to each well along with 10 µl 2XYPD medium. The mating plates
were placed into a resealable plastic bag and incubated for 12-36 hr at 30
°C. Each well was then twice serially-diluted using SGR-UHWL (minimal
selective dropout media minus uracil, histidine, tryptophan and leucine, +
2% galactose, + 1% raffinose) to a final volume of 110 µl. The diluted matings
were incubated for an additional 4-10 days at 30 °C. Ten µl of the mating
mixture were then transferred to a second set of microtiter plates prior to
performing the ß-galactosidase analysis. These mated and out-grown 10
µl stocks were later used to allow recovery of DNA from interacting clones.
Growth of yeast in 96-well plates was measured by absorbance at 600nm
on a Spectramax plate reader using Softmax Pro software.
ß-Galactosidase Assay
Cells were lysed by the addition of 100 µl of a solution of Z buffer [Na2HPO4,
(16.1 g/l), NaH2PO4, (5.5 g/l), KCl (0.75 g/l), and MgSO4, (0.25 g/l), adjusted
to pH 7.0] containing oxalyticase (Enzogenetics) (100 U/ml), 0.1% sodium
dodecyl sulfate (SDS, Sigma), and 2 mg/ml chlorophenyl red-ß-D-
galactopyranoside (CPRG, Boehringer Mannheim). The plates were
incubated at room temperature until sufficient conversion of the CPRG into
its 574nm-absorbing hydrolysis product had occurred (usually 10 min to 2
hr). To quantify the extent of the reaction, the cell debris was removed by
filtration through multiscreen plates (Millipore) and the chlorophenyl red
measured at an absorbance of 574 nm on a Spectramax plate reader using
Softmax Pro software (Molecular Devices Corp.).
Yeast Strains, Plasmids and Media
Yeast strains EGY48 (Matα, ura3, his3, trp1, leu2::6lexAop:LEU2) and
RFY206 (Mata his3∆200 leu2-3 lys2∆201 ura3-52 trp1::hisG) were from R.
Brent. pSH1834T, pRPB7, pRPB4, pEGKRev1, pEGRas, pJGKrit1 and
pJGRaf (Serebriiskii et al., 1997) were gifts of E. Golemis. pEG202 and
pJG45 were from R. Brent. pRS424 was from ATCC. pEG-LXRα, pEG-
RXRα, pJG-LXRα, and pJG-RXRα were constructed by cloning the ligand
binding domains of the nuclear hormone receptors LXRα and RXRα into
pEG202 and pJG45. Complete yeast media (YPD) consists of 1% yeast
extract, 2% peptone, 2% dextrose. Selective yeast media are as described
in Gyuris et al., 1993.
Acknowledgements
The authors wish to thank Mike Watson for pEGLXRα and for technical
assistance throughout this work, the Core Sequencing Group for sequencing
baits and preys, and the members of the Y2H team for their valuable input.
References
Bartel, P.L., Roecklein, J.A., SenGupta, D., and Fields, S. 1996. A protein
linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12: 72-
77.
Bendixen, C., Gangloff, S., and Rothstein, R. 1994.Ayeast mating-selection
scheme for detection of protein-protein interactions. Nucleic Acids Res
22: 1778-1779.
Cho, R.J., Fromont-Racine, M., Wodicka, L., Feierbach, B., Stearns, T.,
Legrain, P., Lockhart, D.J., and Davis R.W. 1998. Parallel analysis of
genetic selections using whole genome oligonucleotide arrays. Proc. Natl.
Acad. Sci. USA. 95: 3752-3757.
Fields, S., and Song, O.K. 1989. A novel genetic system to detect protein-
protein interactions. Nature. 340: 245-246.
Finley, R.L. Jr., and Brent, R. 1994. Interaction mating reveals binary and
ternary connections between Drosophila cell cycle regulators. Proc. Natl.
Acad. Sci. USA. 91: 12980-12984.
Fromont-Racine, M., Rain, J.-C., and Legrain, P. 1997. Toward a functional
analysis of the yeast genome through exhaustive two-hybrid screens.
Nat. Genet. 16: 277-282.
Golemis, E.A., Serebriiskii, I., and Law, S.F. 1997.Adjustment of parameters
in the yeast two-hybrid system: criteria for detecting physiologically
significant protein-protein interactions. Curr. Innovations Mol. Biol. 4: 11-
28.
Gyuris, J., Golemis, E., Chertkov, H., and Brent, R. 1993. Cdi1, a human
G1 and S phase protein phosphatase that associated with Cdk2. Cell.
75: 791-803.
Hengen, P.N. 1997. False positives from the yeast two-hybrid system. Trends
Biochem. Sci. 22: 33-34.
Janowski, B.A., Willy, P.J., Devi, T.R., Falck, J.R., and Mangelsdorf, D.J.
1996. An oxysterol signalling pathway mediated by the nuclear receptor
LXR alpha. Nature. 383: 728-731.
Le Douarin, B., Nielsen, A.L., Garnier, J.M., Ichinose, H., Jeanmougin, F.,
Losson, R., and Chambon, P.A. 1996. Possible involvement of TIF1alpha
and TIF1beta in the epigenetic control of transcription by nuclear receptors.
EMBO J. 15: 6701-6715.
Lee, J.W., Choi, H.S., Gyuris, J., Brent, R., and Moore, D.D. 1995. Two
classes of proteins dependent on either the presence or absence of thyroid
hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol.
9: 243-254.
McCormack, A.L., Schieltz, D.M., Goode, B., Yang, S., Barnes, G., Drubin,
D., and Yates, J.R., III. 1997. Direct analysis and identification of proteins
in mixtures by LC/MS/MS and database searching at the low-femtomole
level. Anal. Chem. 69: 767-776.
McKune, K., Richards, K.L., Edwards, A.M., Young, R.A., and Woychik,
N.A. 1993. RPB7, one of two dissociable subunits of yeast RNA
polymerase II, is essential for cell viability. Yeast. 9: 295-299.
Nagase, T, Seki, N., Ishikawa, K., Ohira, M., Kawarabayasi, Y., Ohara, O.,
Tanaka, A., Kotani, H., Miyajima, N., and Nomura, N. 1996. Prediction of
the coding sequences of unidentified human genes. VI. The coding
sequences of 80 new genes (KIAA0201-KIAA0280) deduced by analysis
of cDNA clones from cell line KG-1 and brain. DNA Res. 3: 321-329.
Paltauf, F., Kohlwein, S.D., and Henry, S.A. 1992. Regulation and
Compartmentalization of Lipid Synthesis in Yeast. In: The Molecular and
Cellular Biology of the Yeast Saccharomyces: Gene Expression, Cold
Spring Harbor Laboratory Press, Plainview, N.Y. p. 415-500.
Serebriiskii, I., Estojak, J., Sonoda, G., Testa, J.R., and Golemis, E.A. 1997.
Association of Krev-1/rap1A with Krit1, a novel ankyrin repeat-containing
protein encoded by a gene mapping to 7q21-22. Oncogene. 15: 1043-
1049.](https://image.slidesharecdn.com/7e7696bb-2ad8-4ac3-8b12-805d7ba1f0e6-160607151127/85/JMMB1_1-5-320.jpg)
