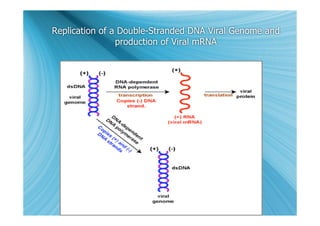
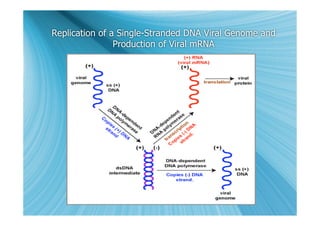
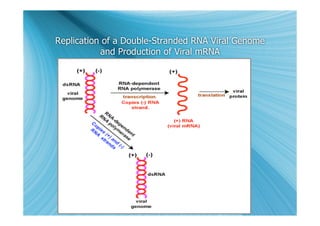
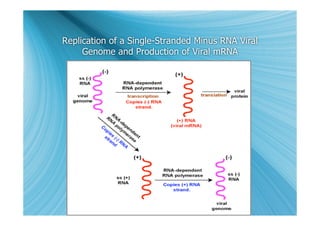
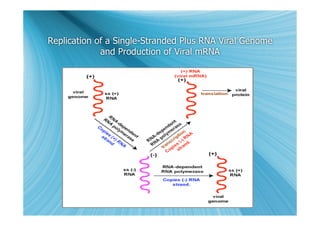
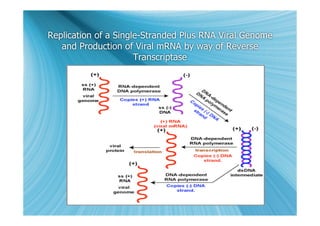
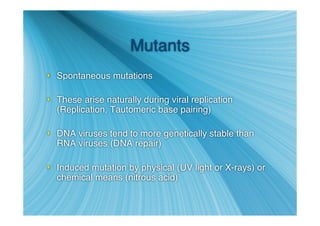
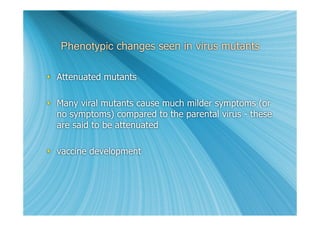
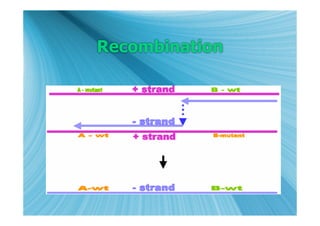
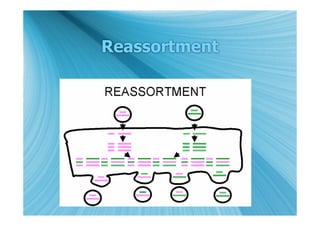
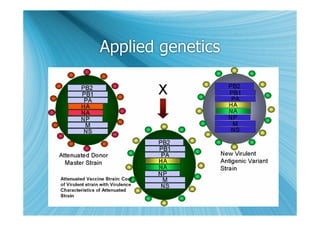
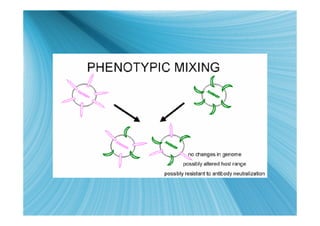
The document discusses the different types of nucleic acids that viruses can use to store their genetic information, including double-stranded DNA, single-stranded DNA, double-stranded RNA, negative-sense RNA, positive-sense RNA, and positive-sense RNA retroviruses. It provides details on how each type replicates and produces viral mRNA.