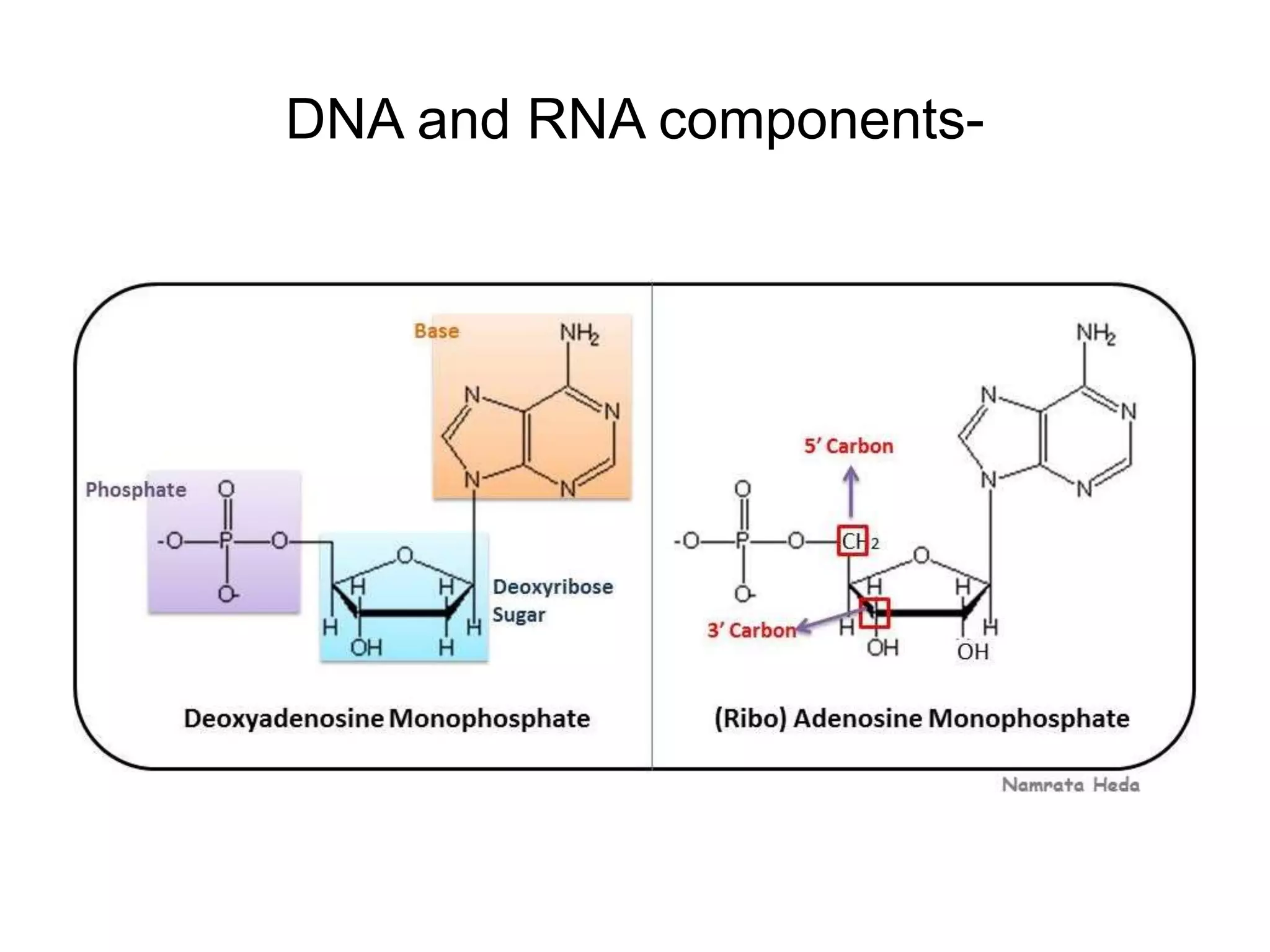
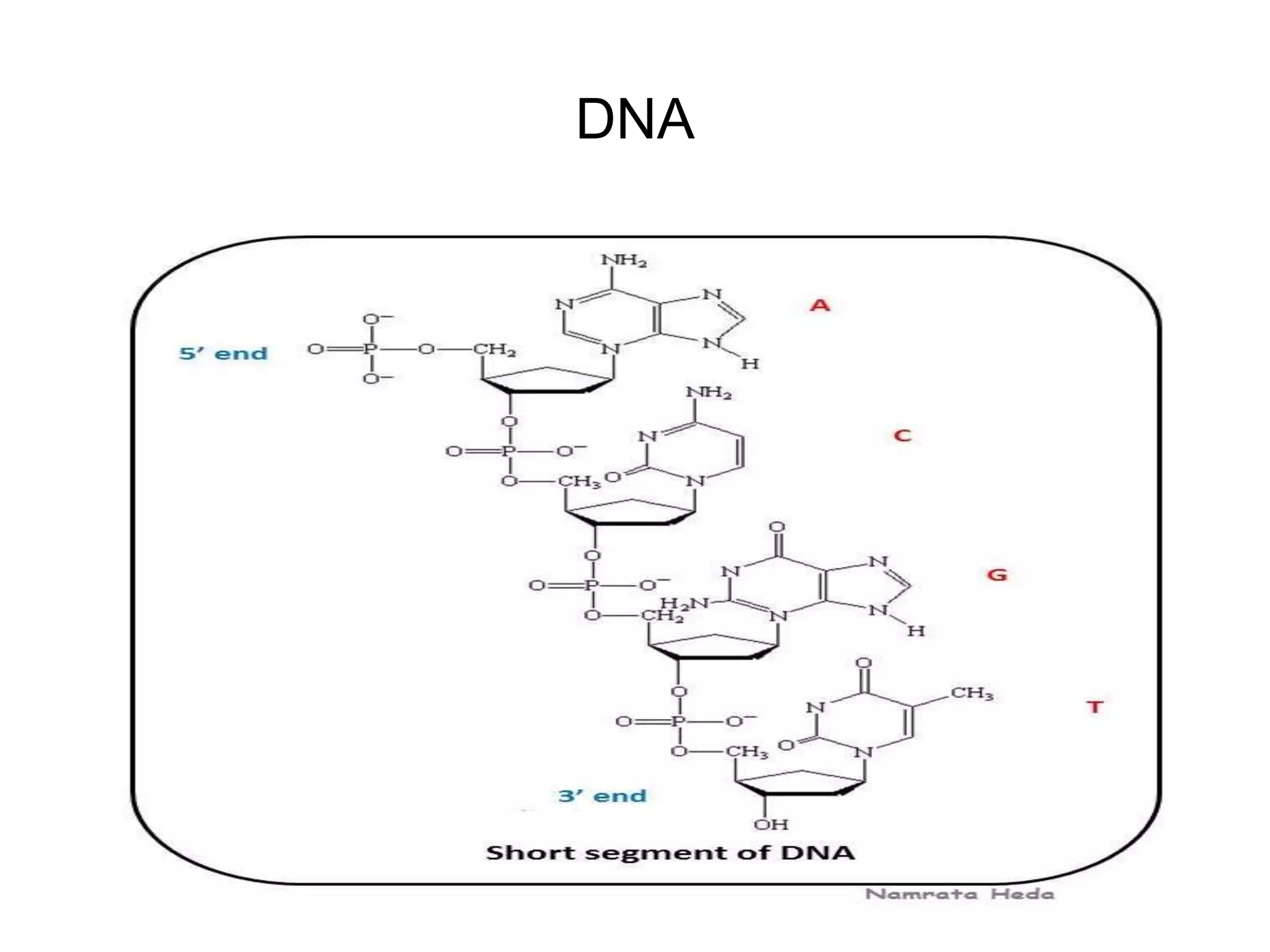
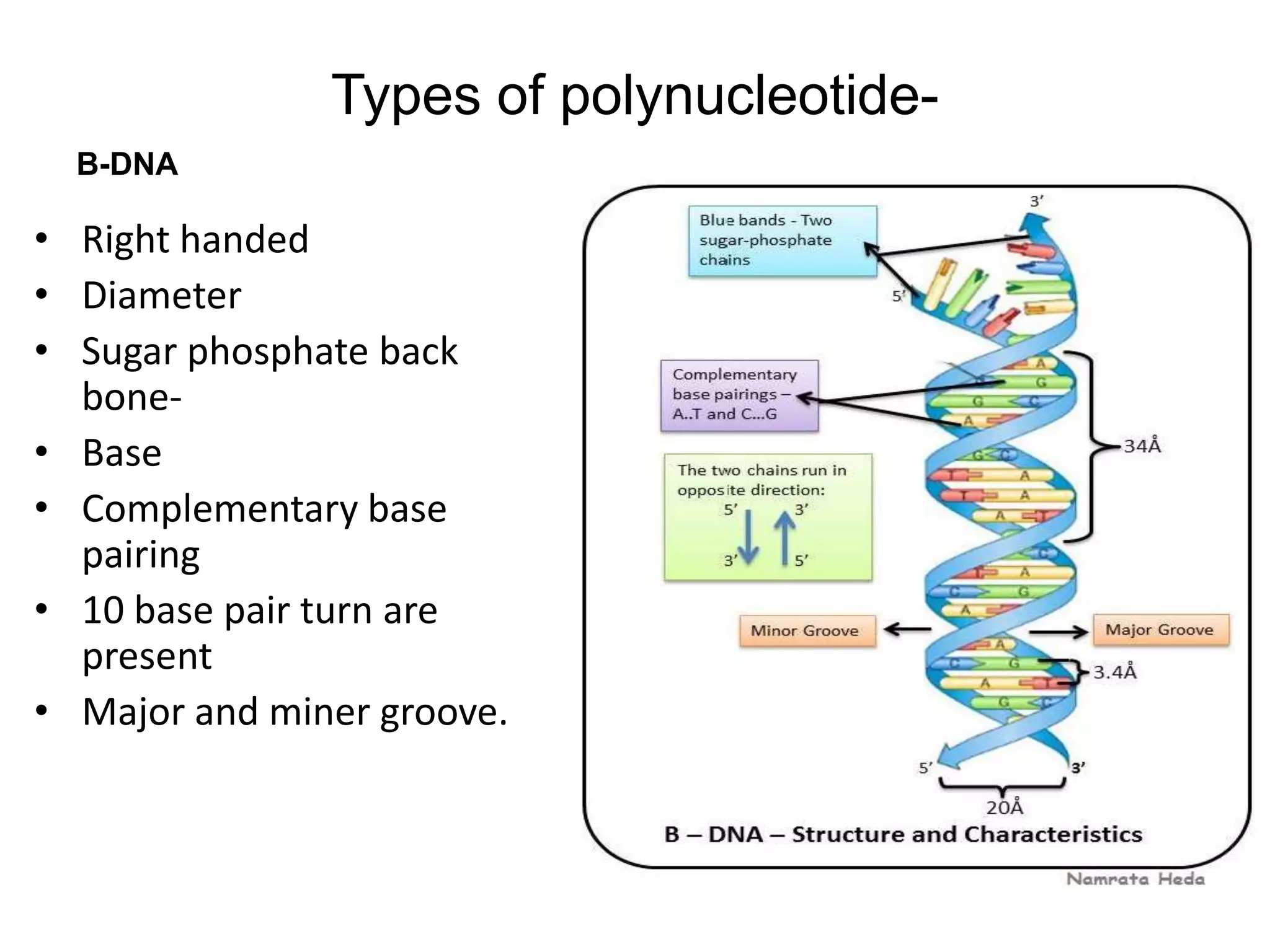
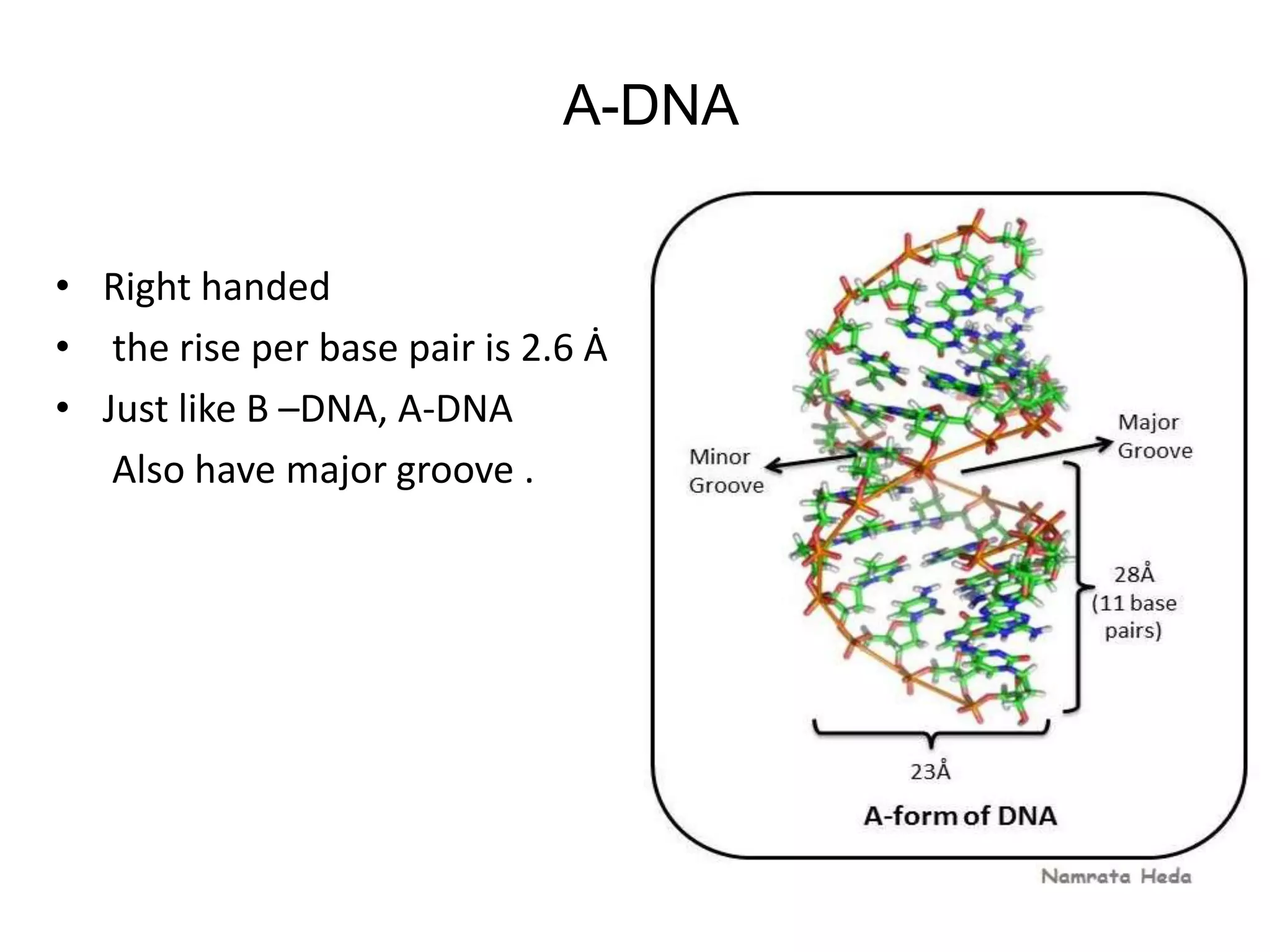
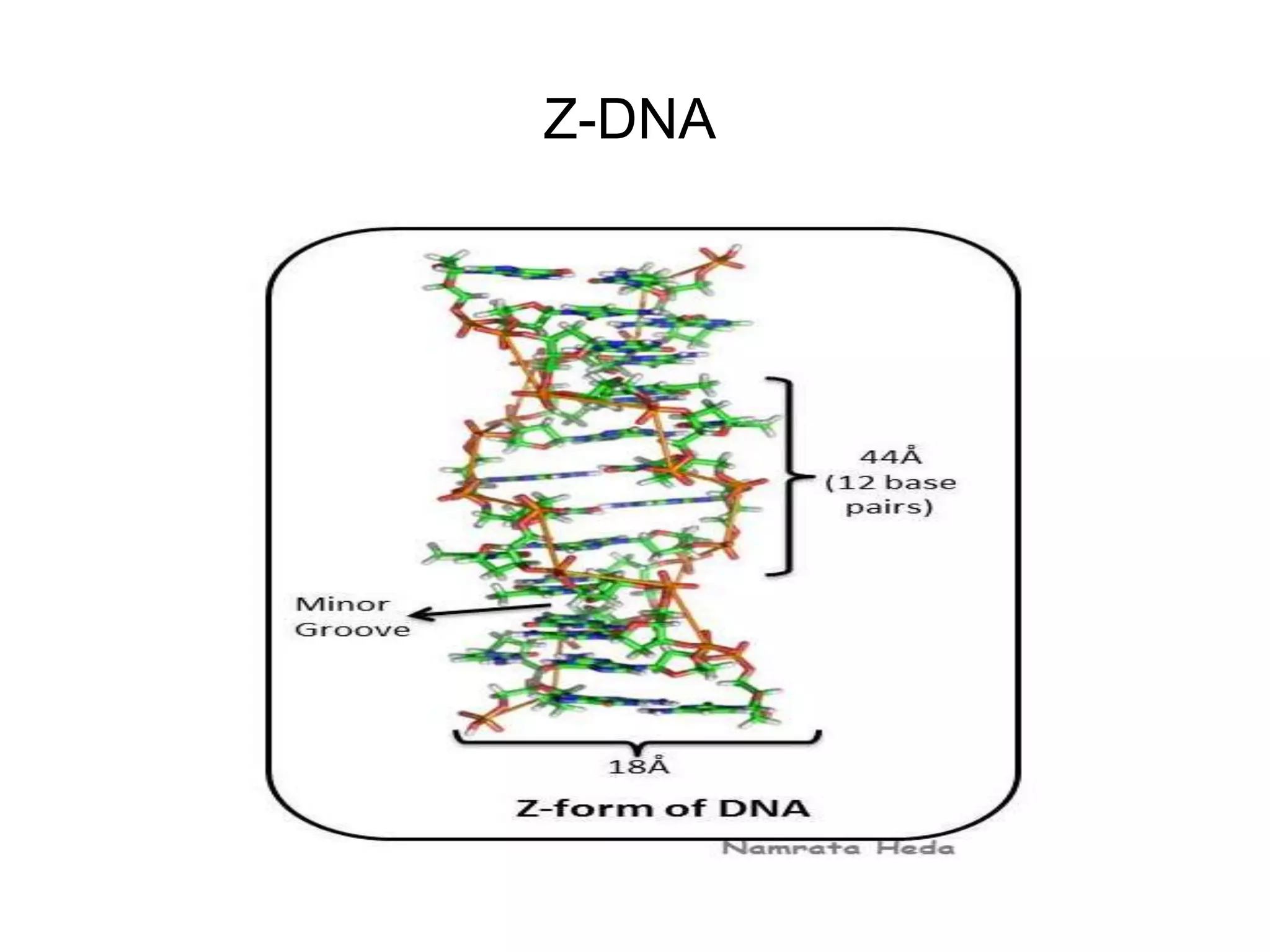
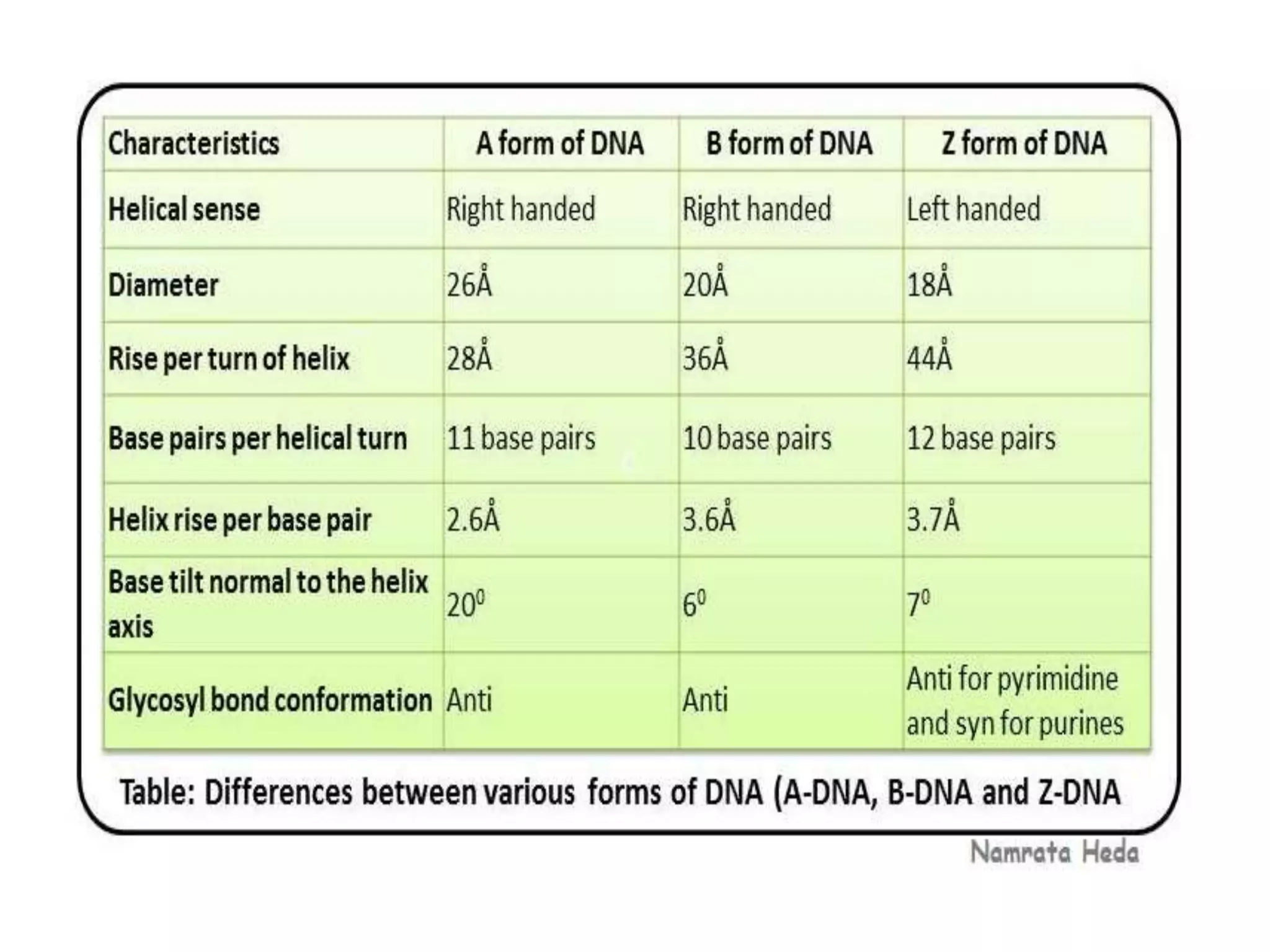
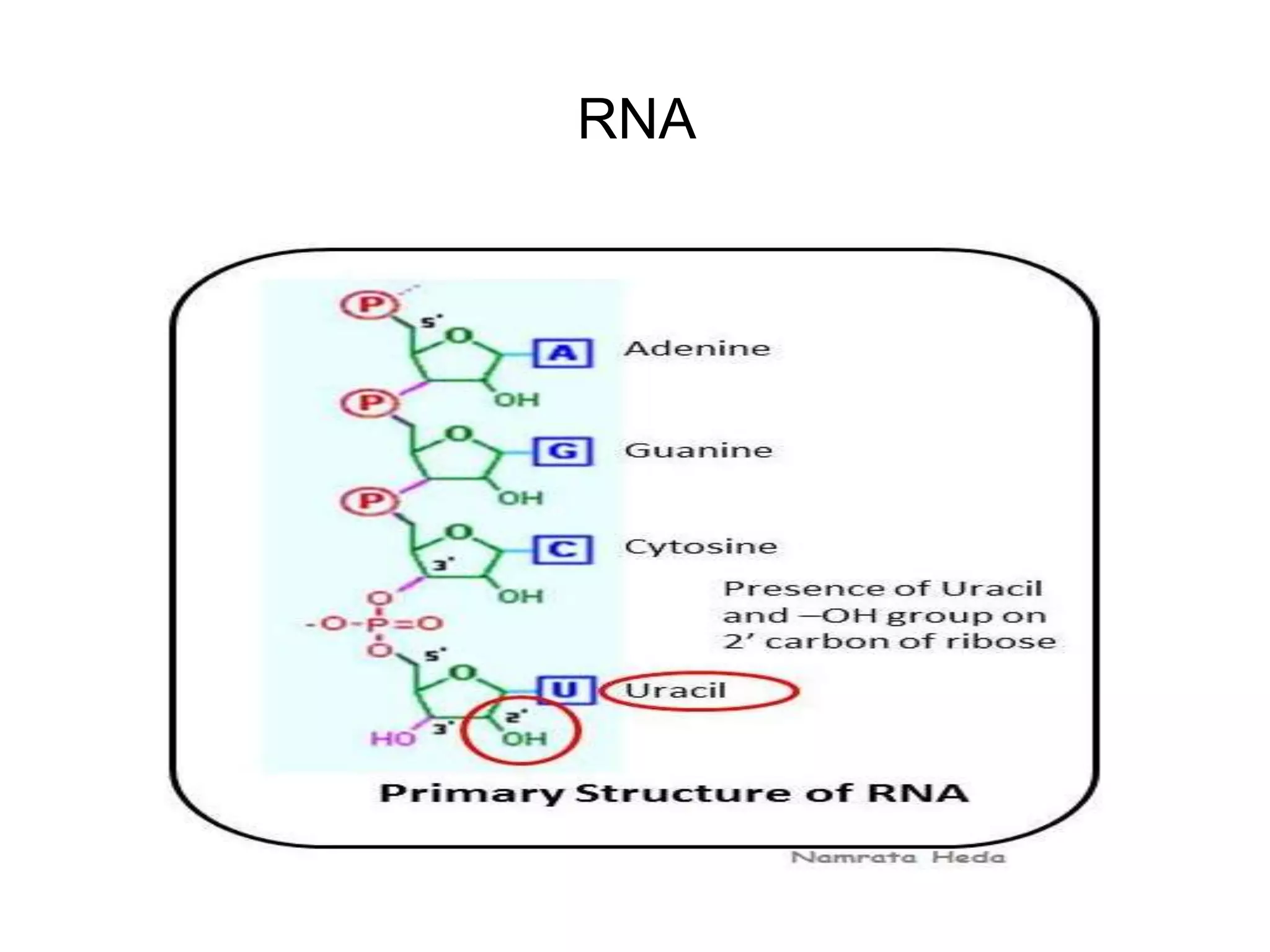
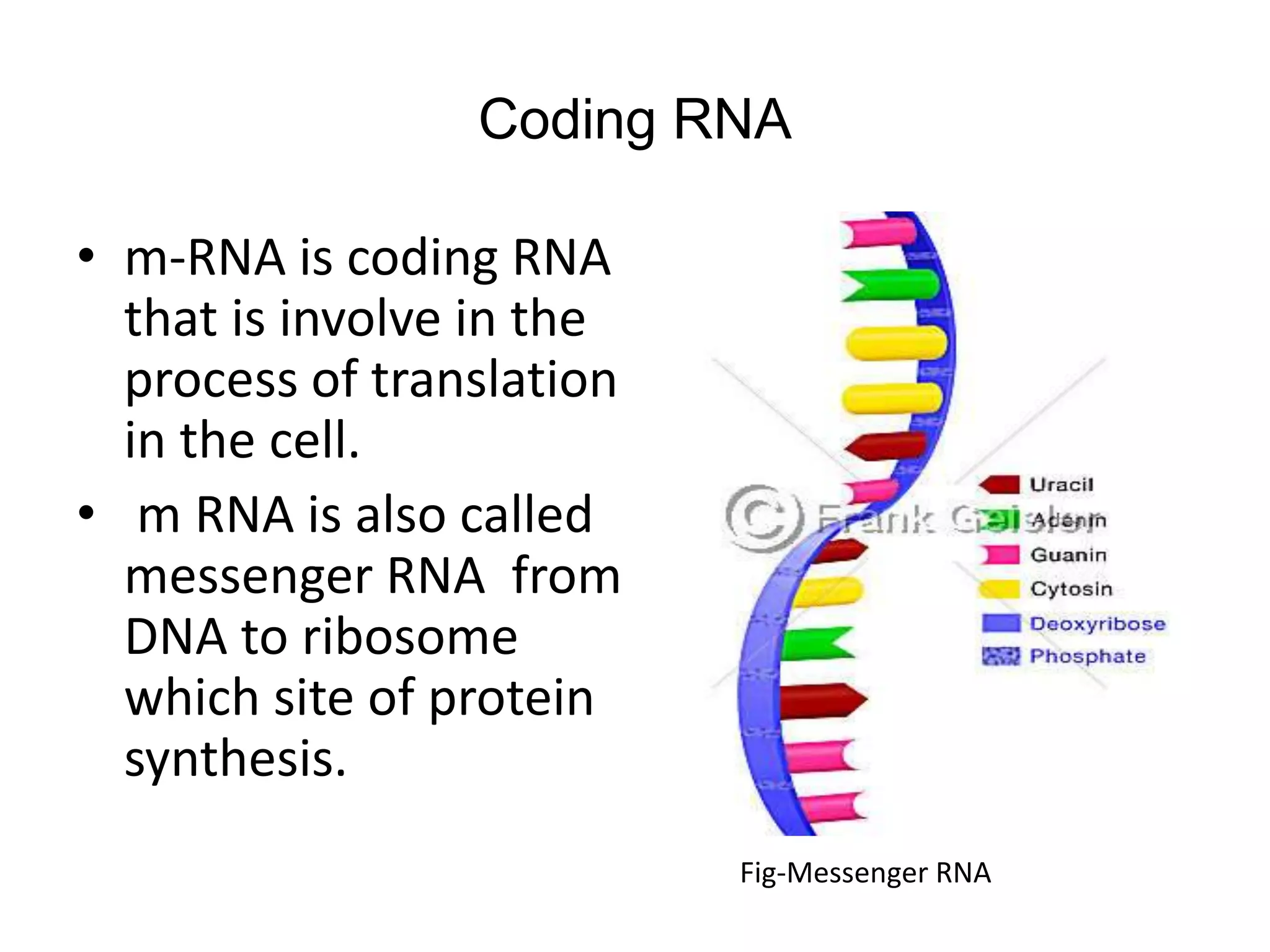
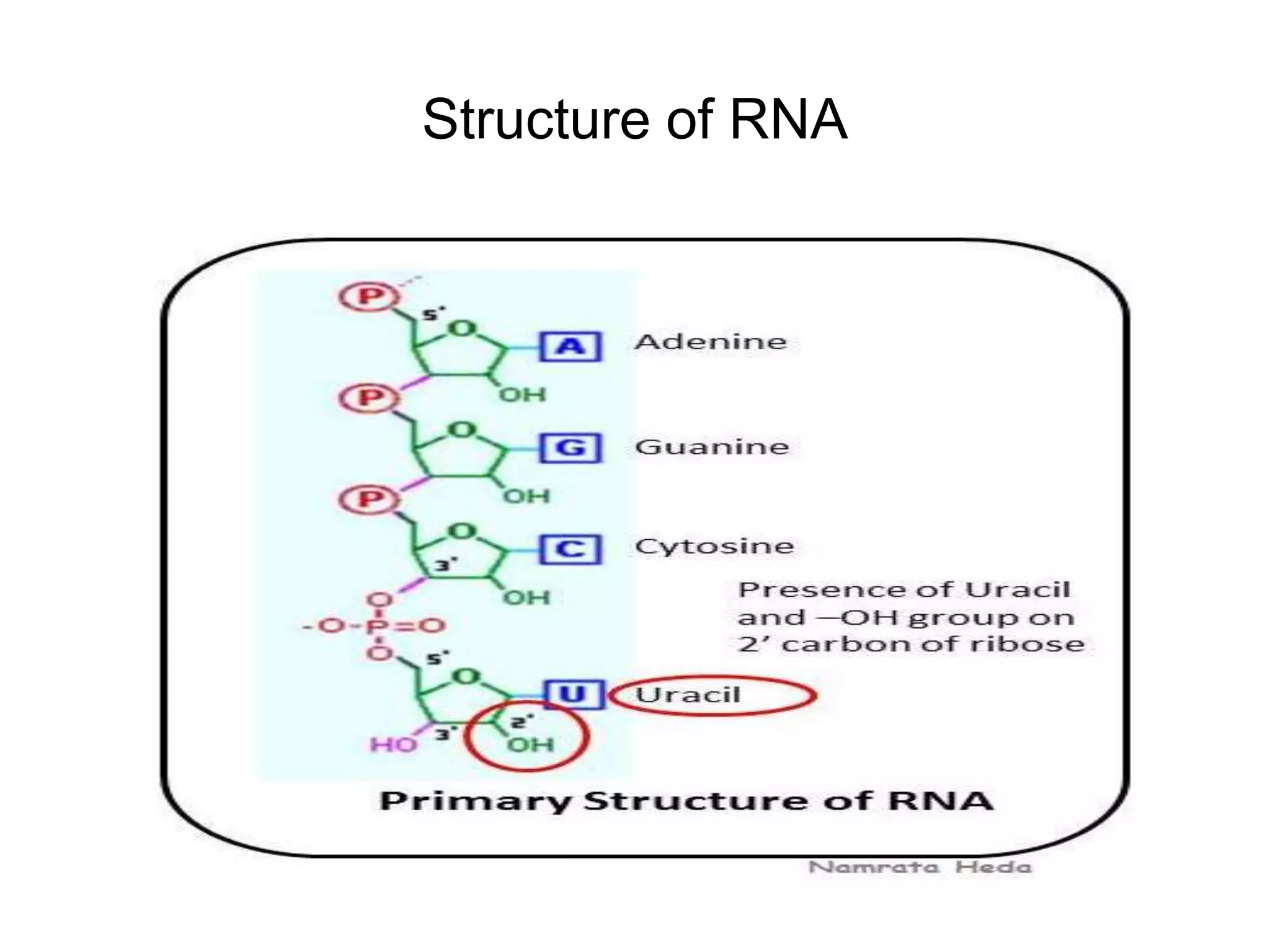
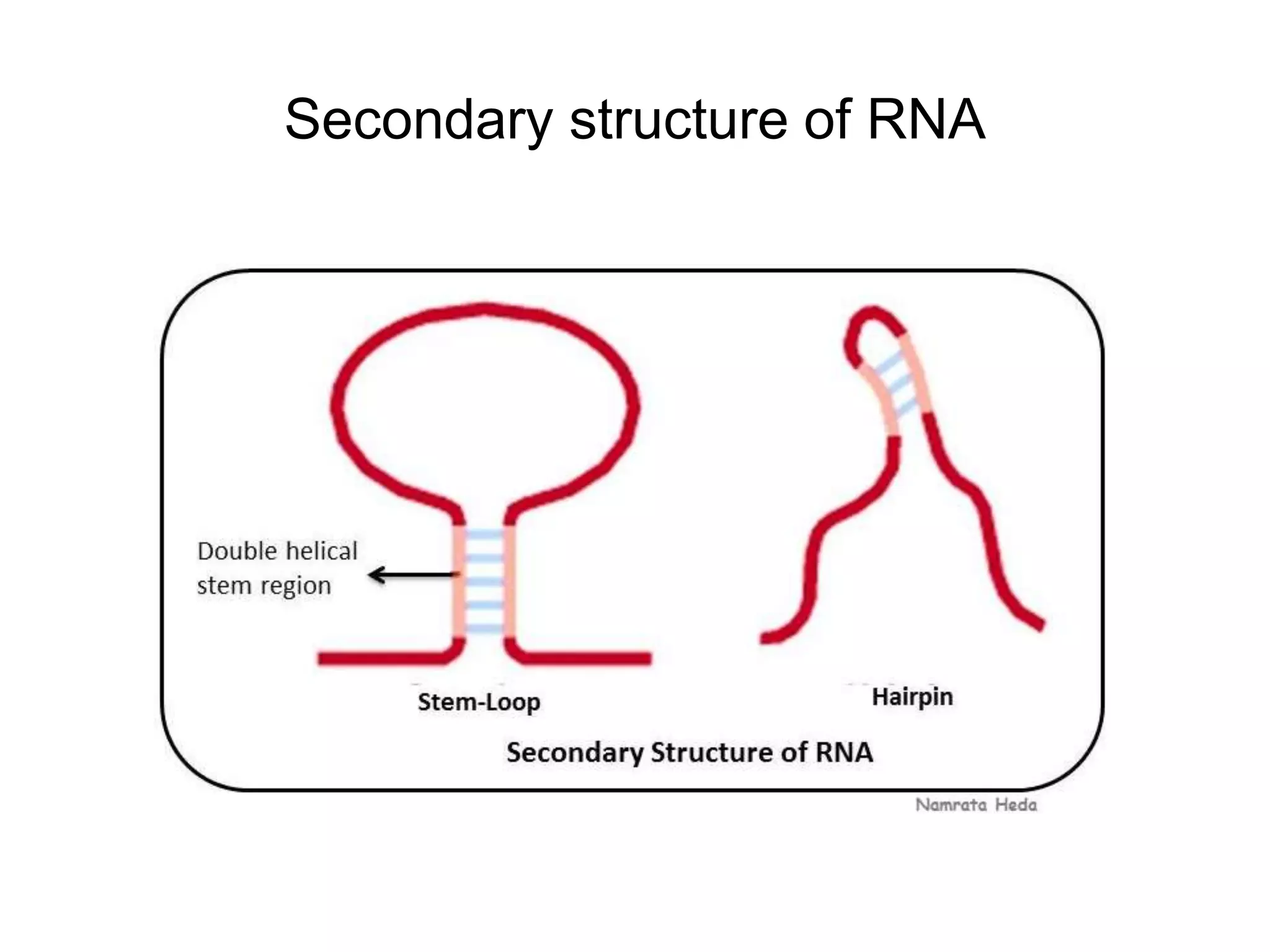
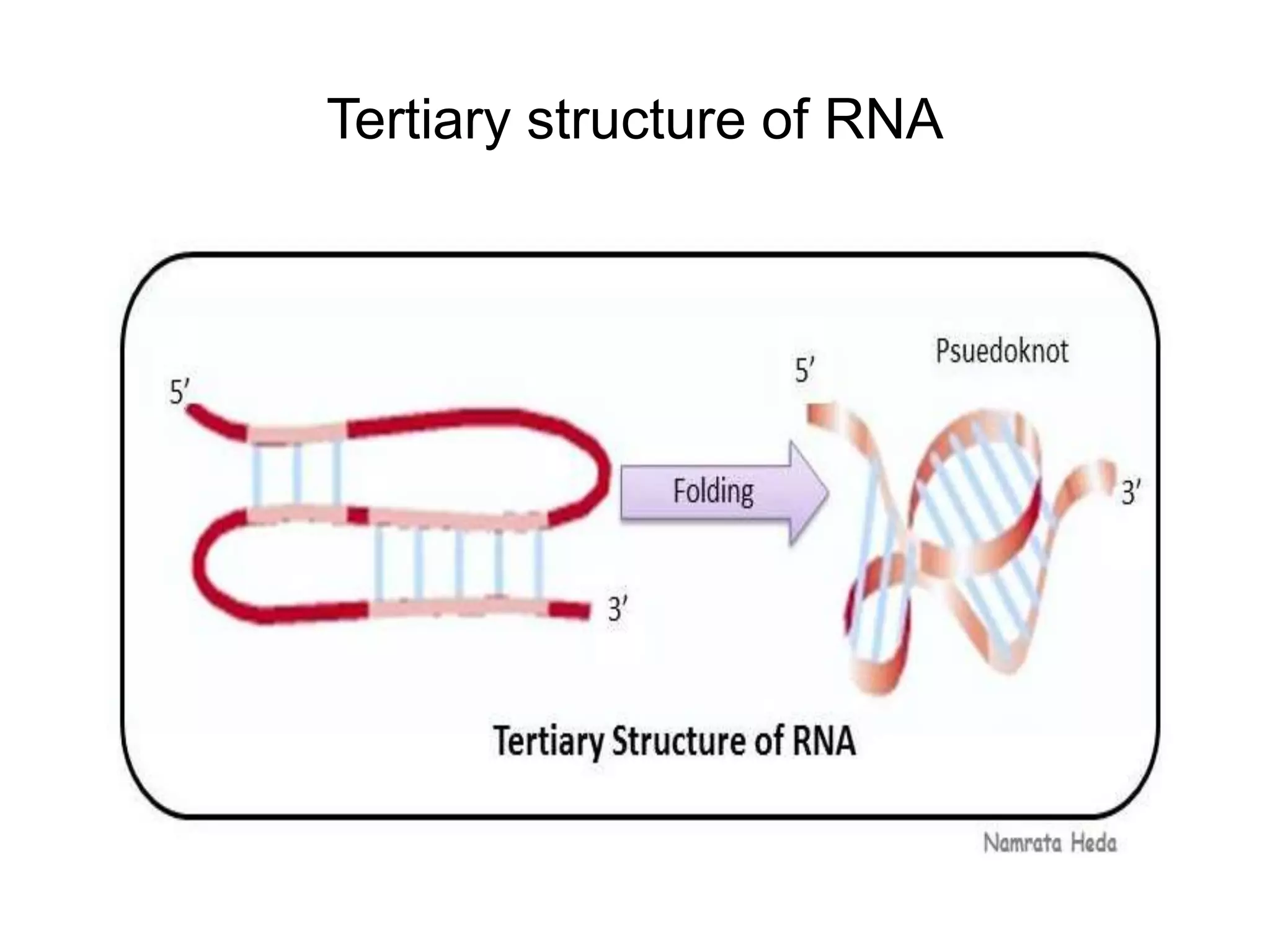
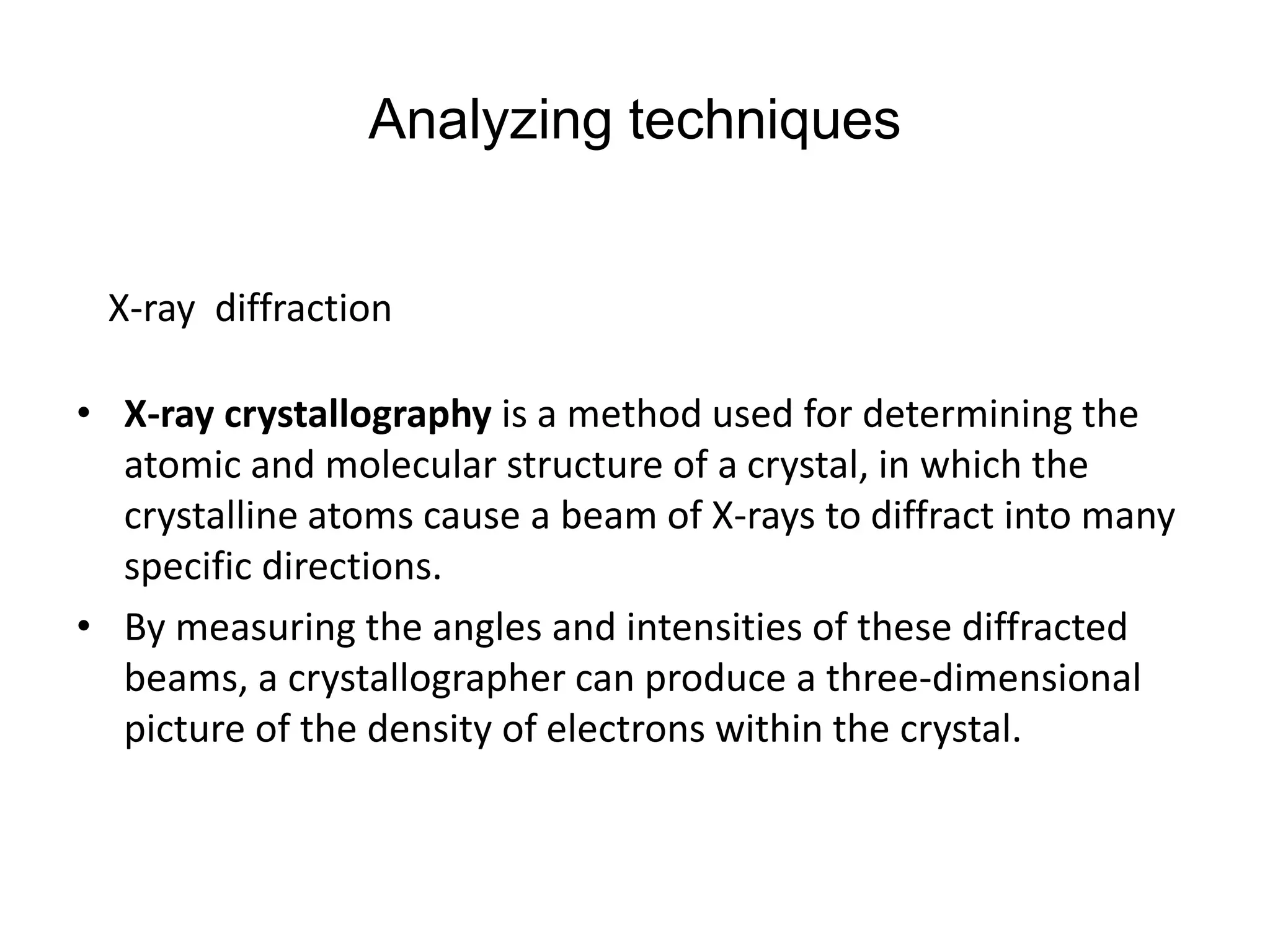
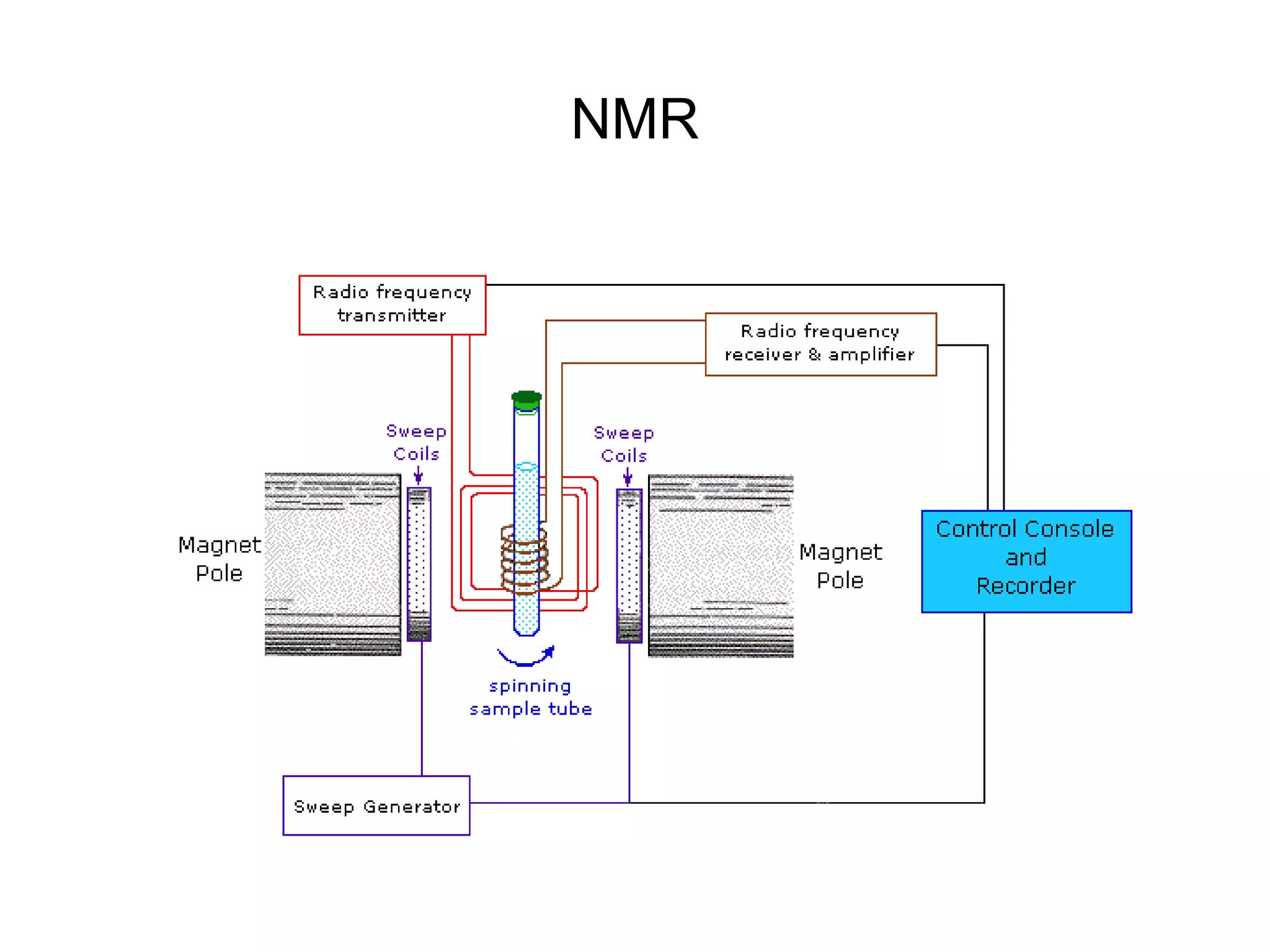
This document discusses the experimental study of polynucleotide conformational properties. It provides an introduction to polynucleotides and their history. It describes the different types of polynucleotides including DNA forms B-DNA, A-DNA, Z-DNA and RNA forms including coding, non-coding, and their primary, secondary, and tertiary structures. The document also discusses techniques used to analyze polynucleotide conformations such as X-ray crystallography, NMR spectroscopy, and spectrophotometry.