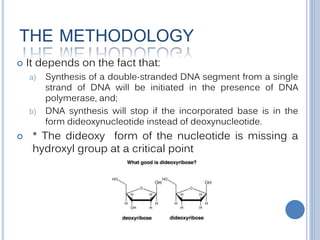
The Sanger technique is a method for DNA sequencing developed in 1977 that uses chain-terminating dideoxynucleotides. It relies on the fact that DNA synthesis will stop if a dideoxynucleotide is incorporated instead of a normal deoxynucleotide. The technique involves incubating a single-stranded DNA with the four dideoxynucleotides separately, which results in DNA fragments of different lengths that can be separated via gel electrophoresis to determine the sequence.