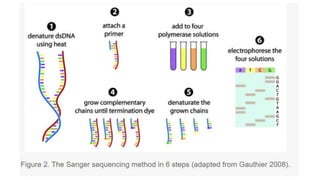
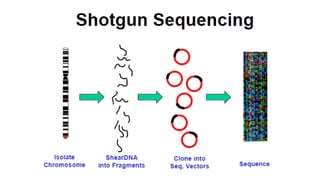
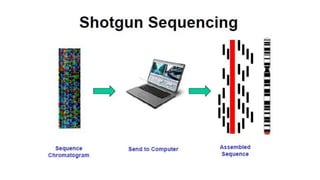
Sanger sequencing, developed by Frederick Sanger in 1977, is a method for determining the nucleotide sequence of DNA, offering 99.99% accuracy, making it the gold standard for validation, especially for small DNA fragments. While next-generation sequencing (NGS) allows for high-throughput analysis of multiple genes and entire genomes, Sanger sequencing is still preferred for cost-efficient single gene sequencing and verification tasks. The process involves denaturing double-stranded DNA, using a primer for DNA synthesis, and employing dideoxynucleotides to terminate sequences for analysis via gel electrophoresis.