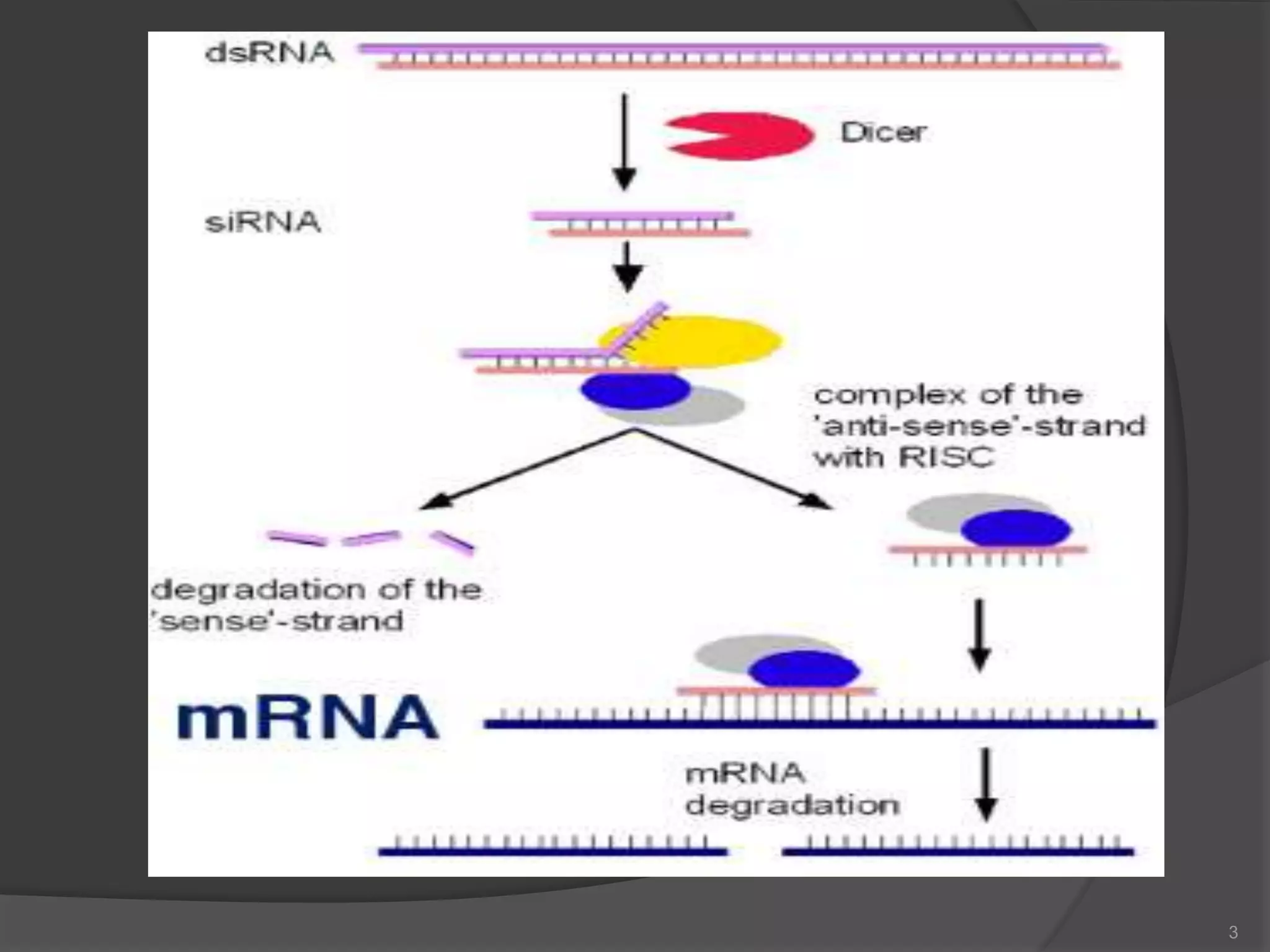
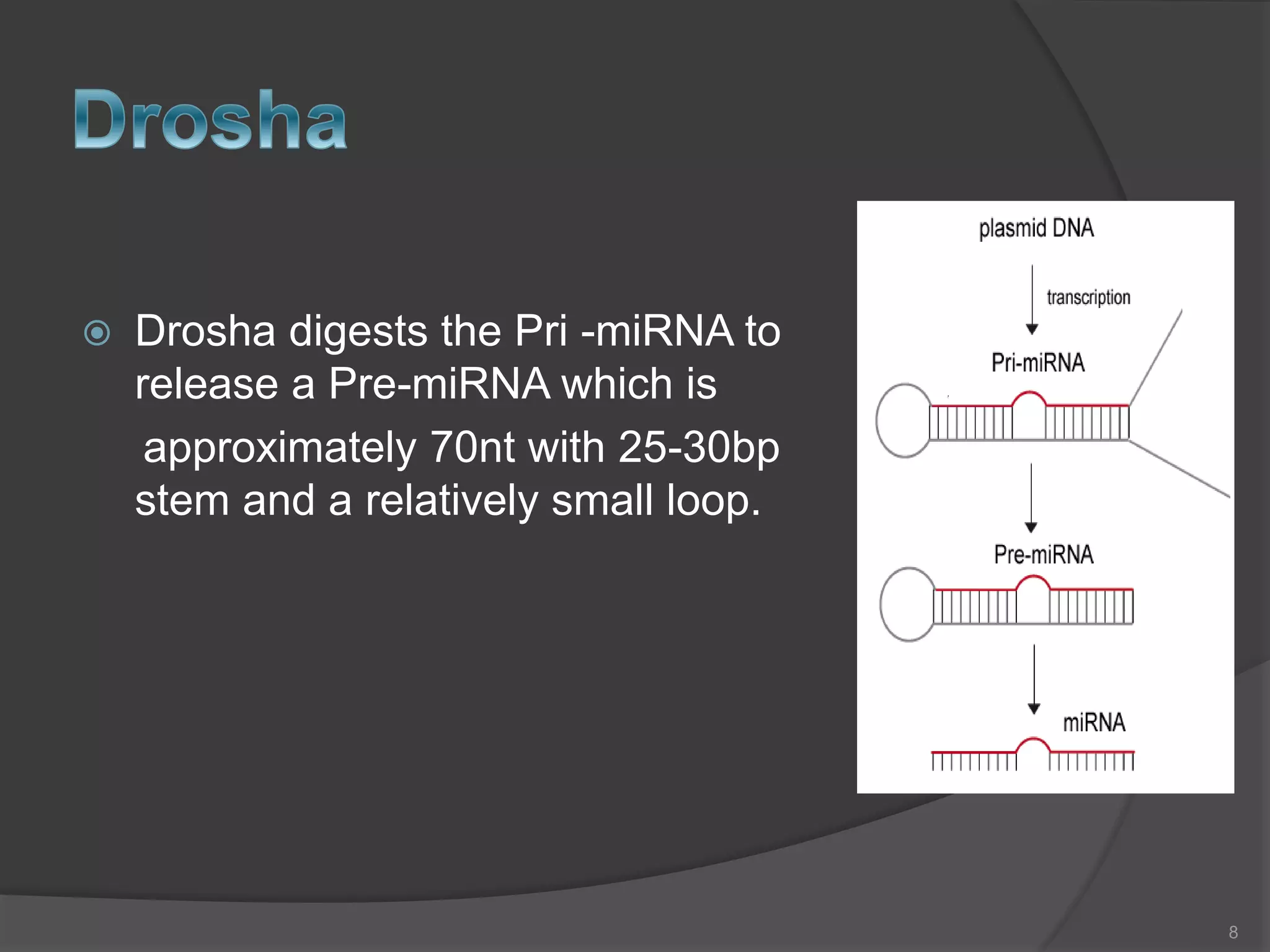
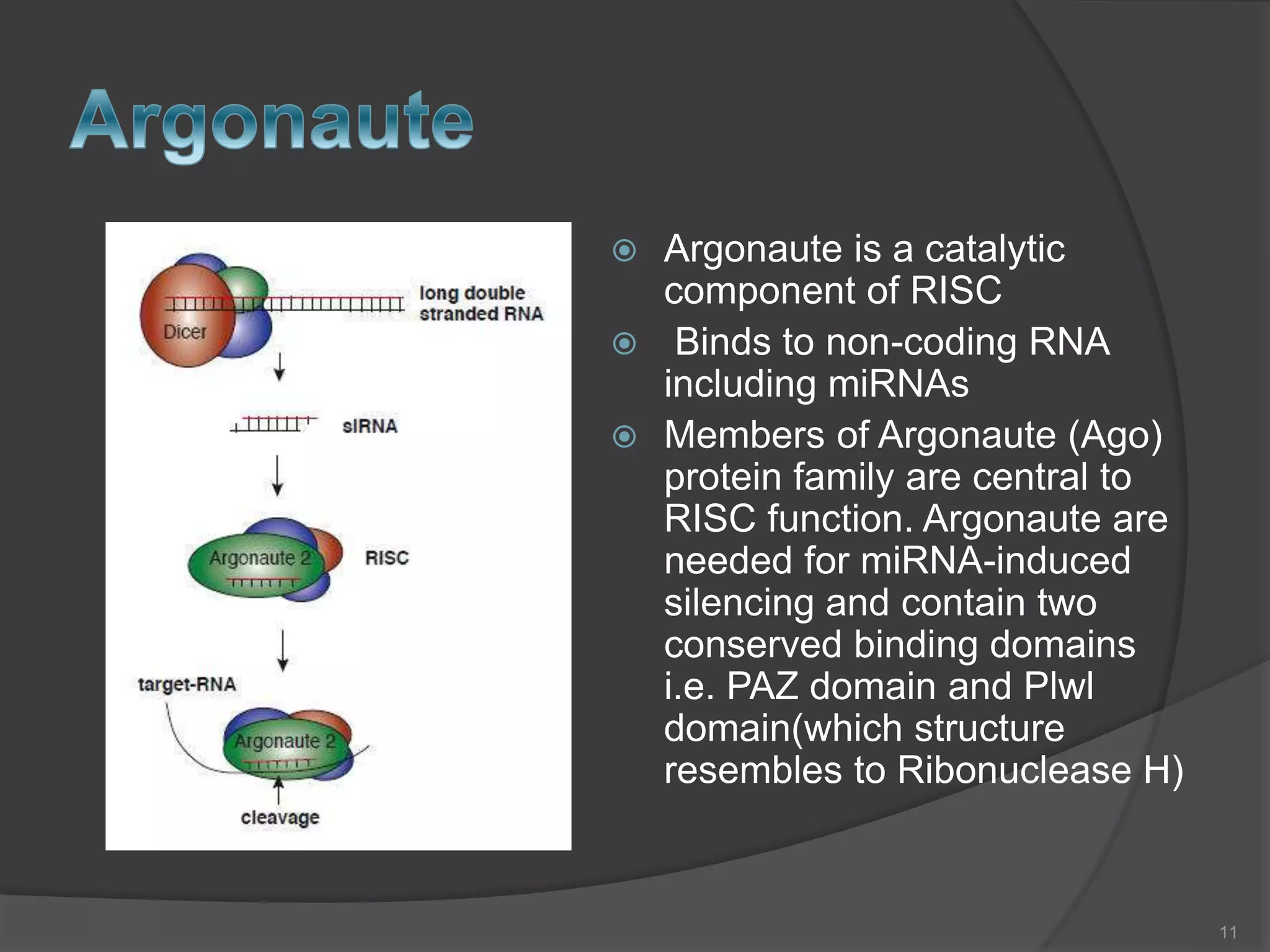
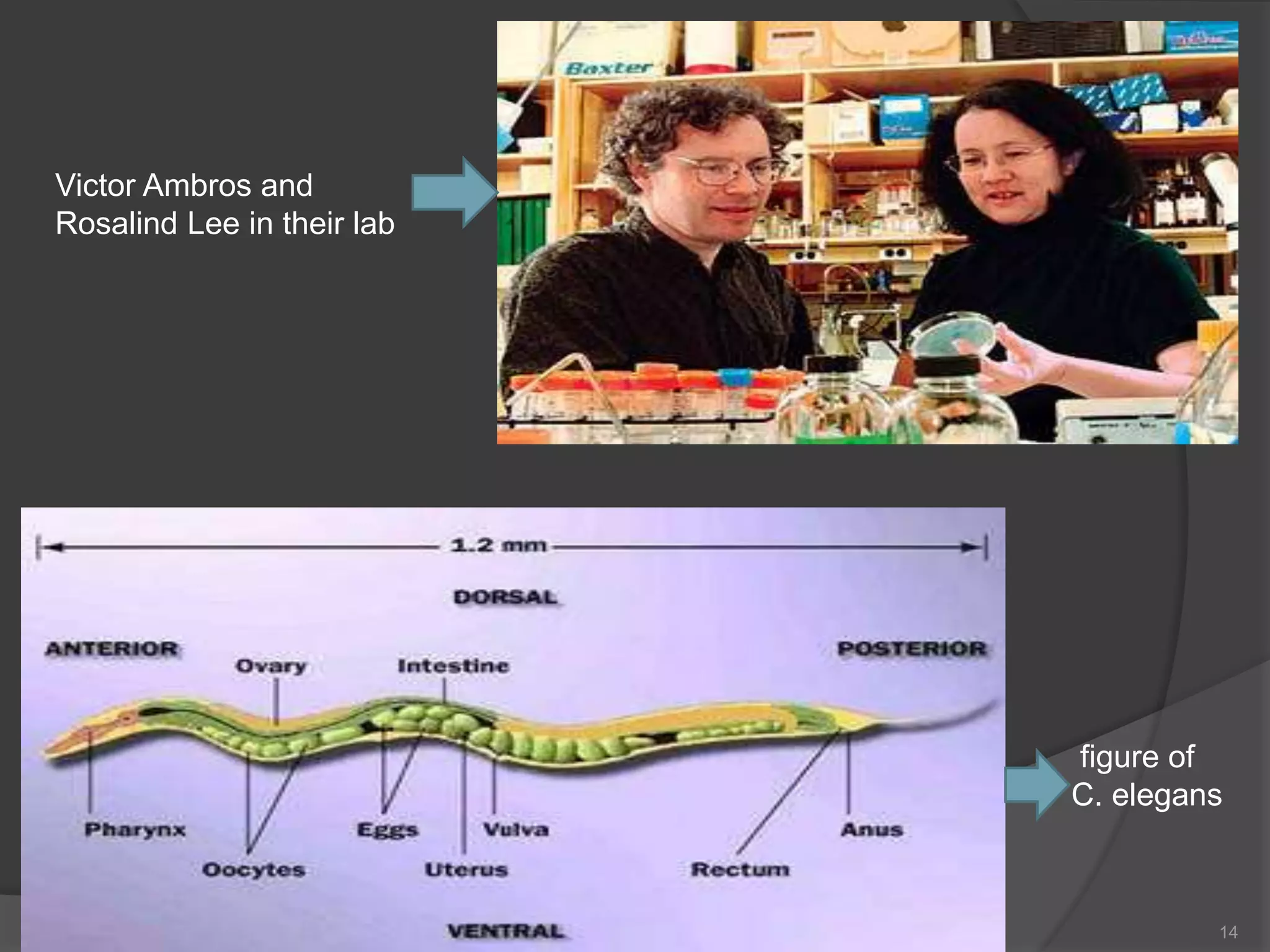
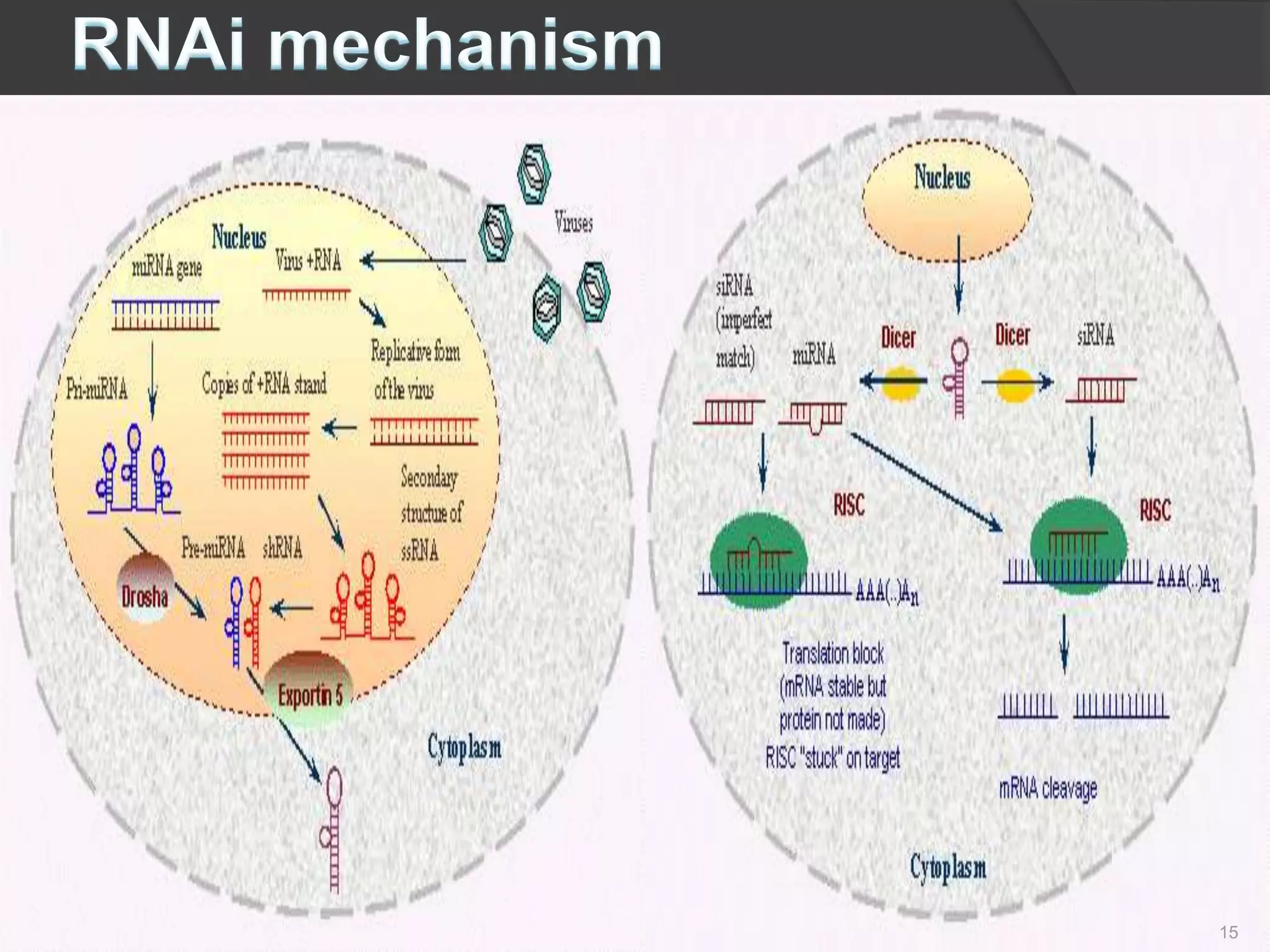
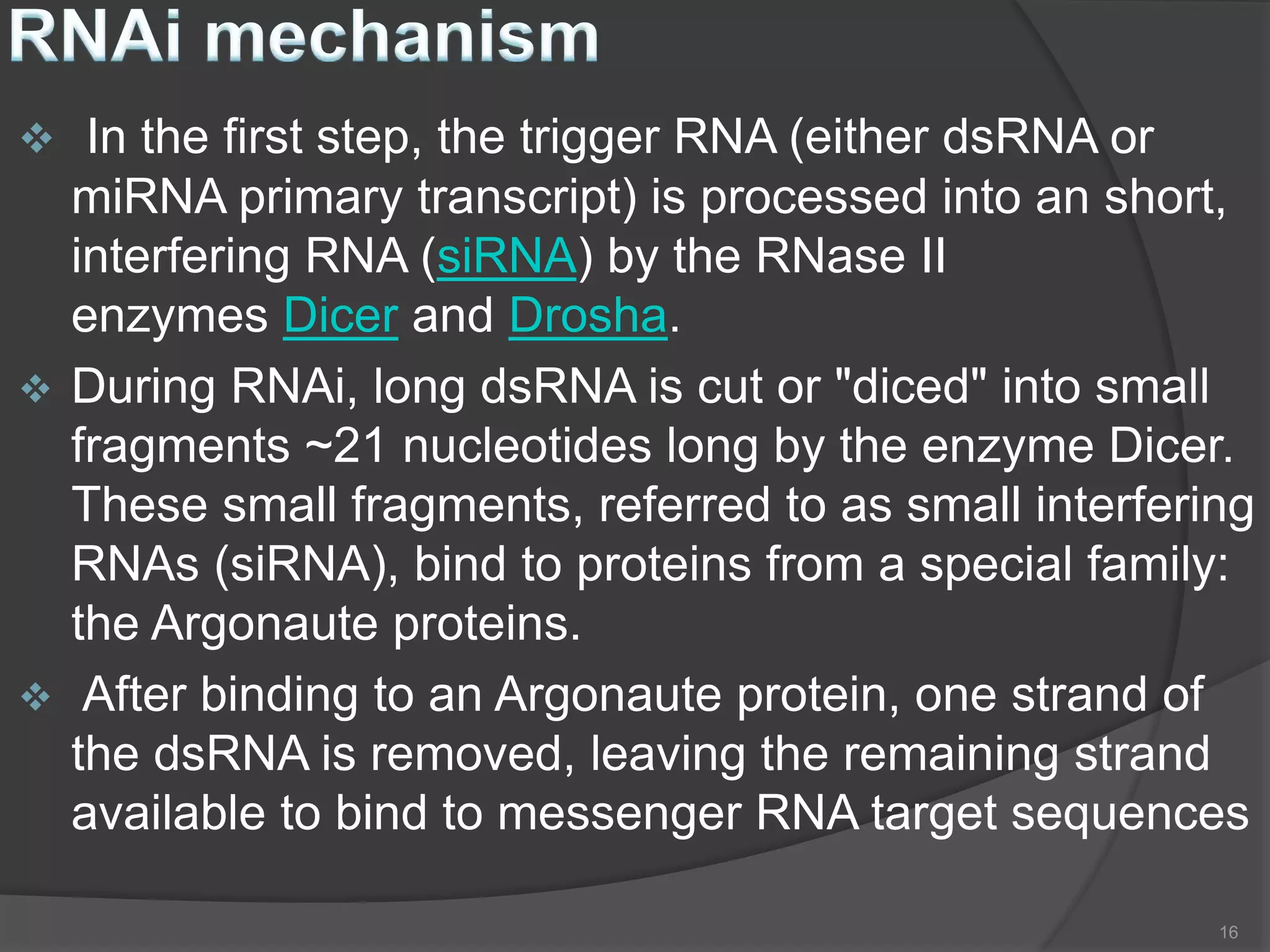
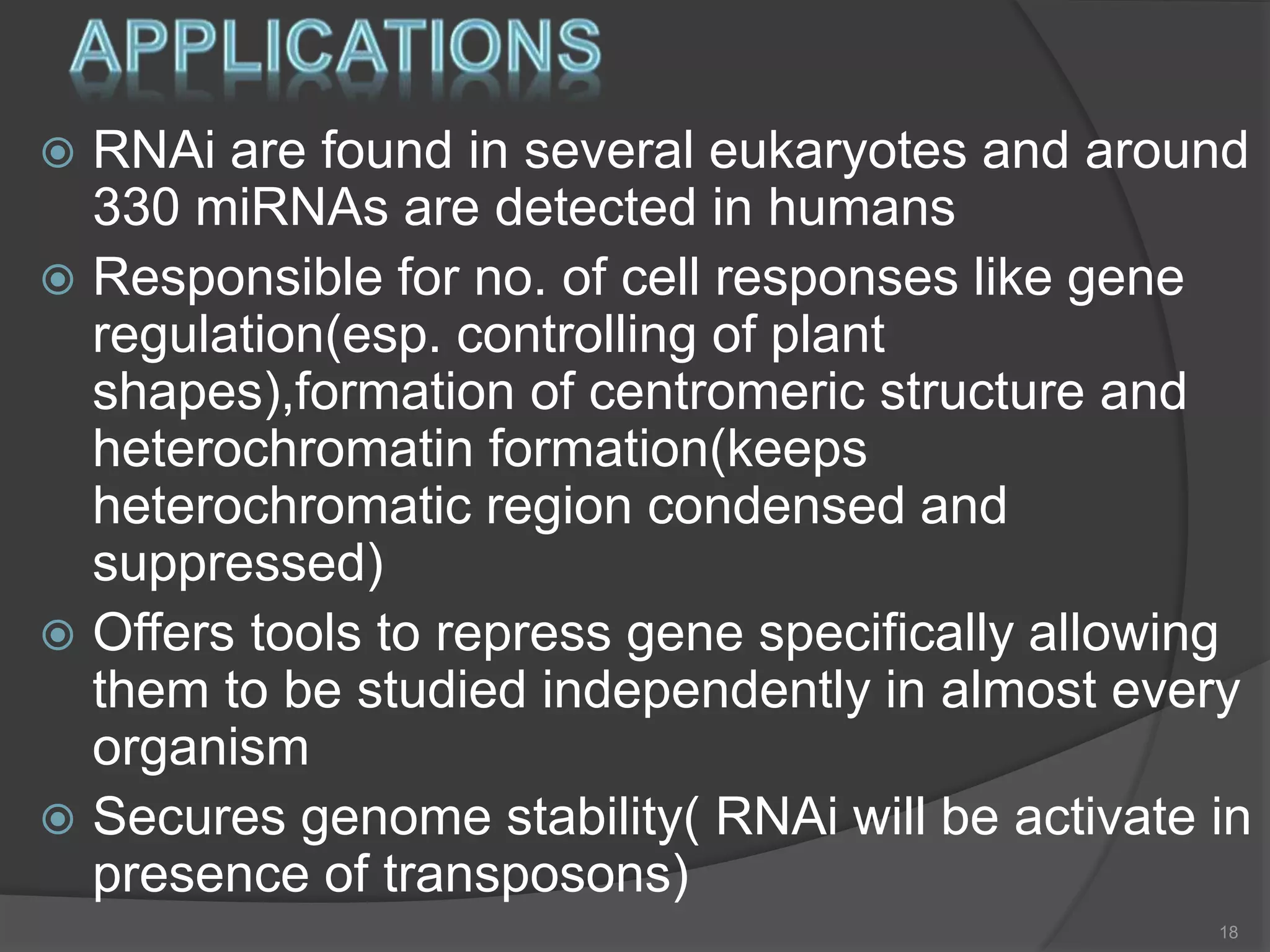
RNA interference is a natural mechanism that inhibits gene expression. It was first discovered accidentally in 1990 when researchers trying to increase pigmentation in petunias found that introduced homologous RNA led to less pigmentation. Further work found similar mechanisms in plants and fungi termed cosuppression and quelling. Craig Mello and Andrew Fire's 1998 paper demonstrating gene silencing in C. elegans using double stranded RNA led to the coining of the term RNAi. The mechanism involves Dicer and Drosha enzymes processing trigger RNA into siRNAs which are loaded into the RISC complex containing Argonaute proteins to degrade complementary mRNA, silencing gene expression. RNAi plays roles in gene regulation, genome stability, and provides therapeutic tools for disease