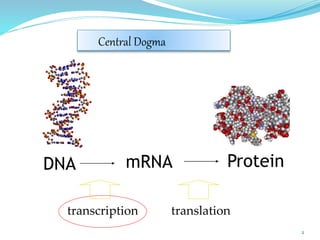
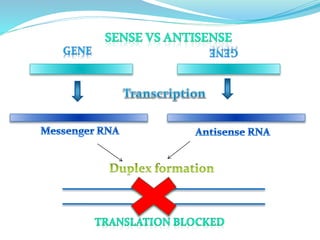
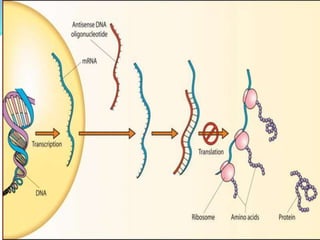
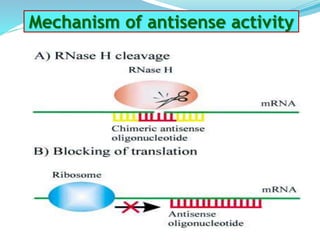
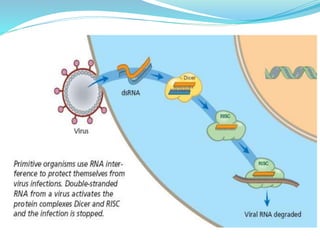
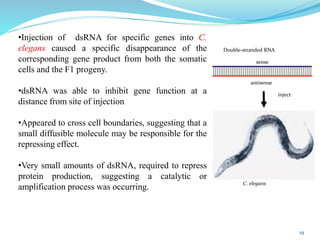
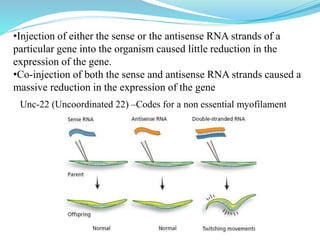
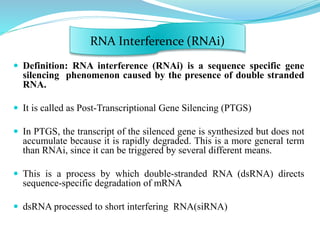
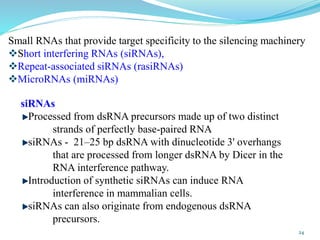
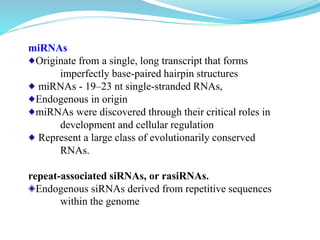
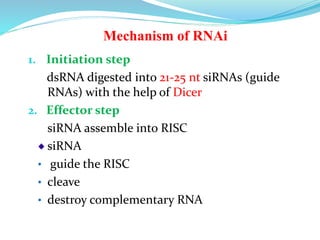
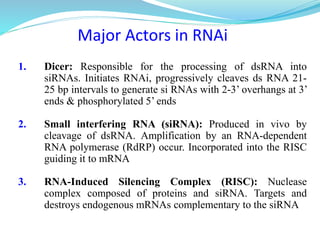
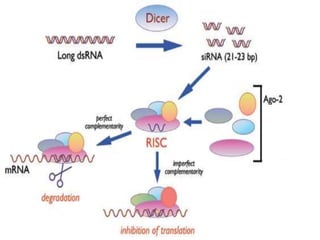
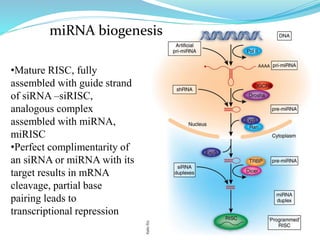
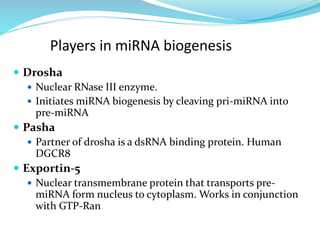
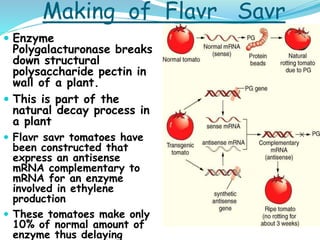
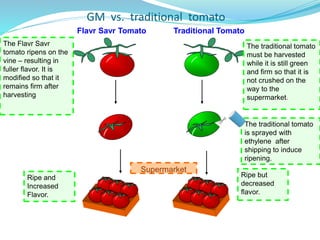
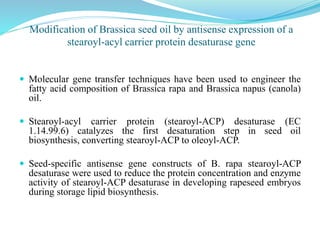
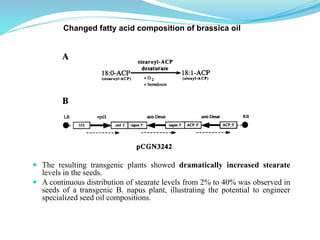
The document discusses antisense RNA technology and its role in gene expression inhibition, detailing its history and mechanisms, including RNA interference (RNAi) and its components like Dicer and RISC. It highlights applications in genetically modified organisms like the Flavr Savr tomato, which utilizes antisense RNA to enhance shelf life and flavor. The technology presents a promising avenue for research and therapeutic development while also raising safety and ethical considerations.