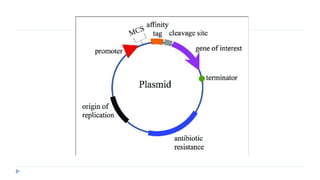
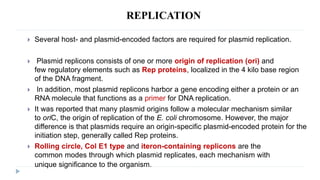
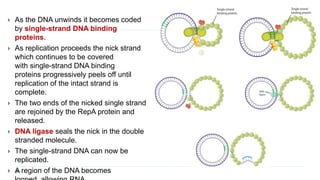
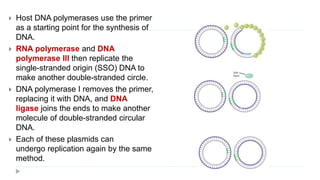
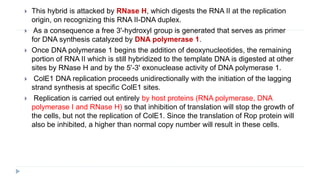
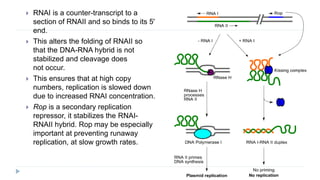
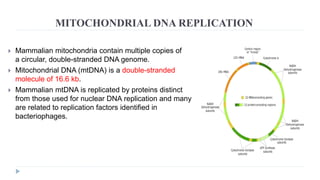
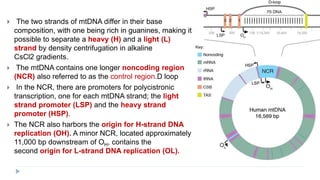
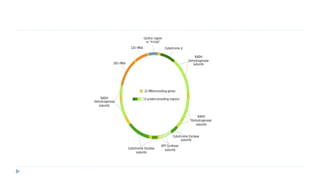
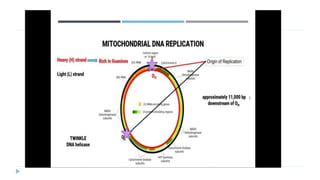
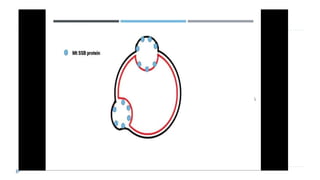
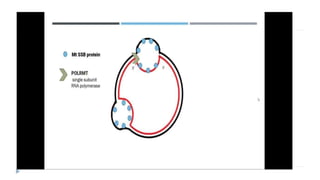
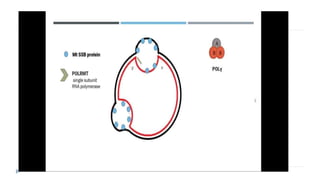
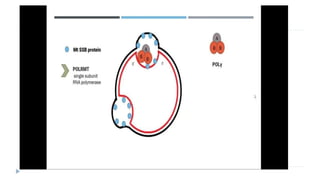
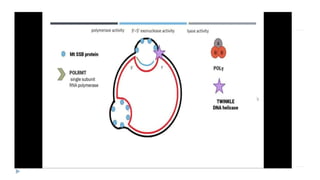
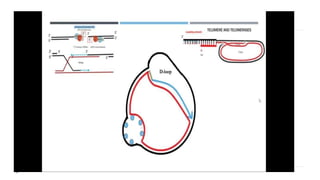
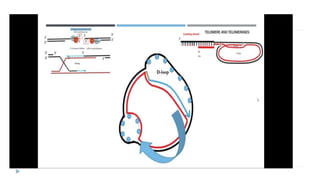
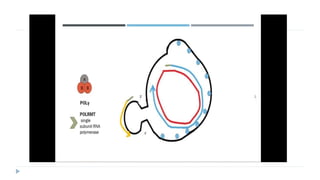
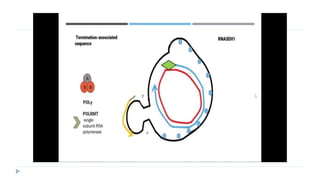
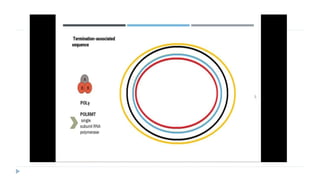
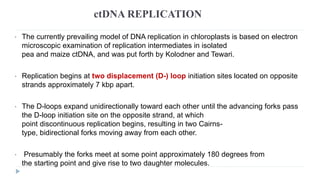
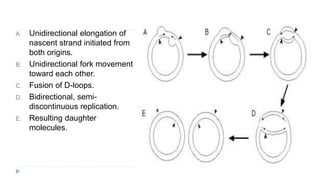
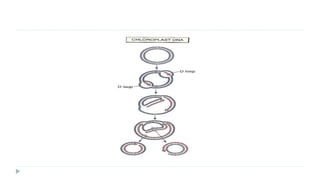
This document discusses extrachromosomal DNA replication in prokaryotes and eukaryotes. In prokaryotes, extrachromosomal DNA is primarily found in plasmids. Plasmids replicate via rolling circle, iteron-regulated, or RNA-regulated mechanisms. In eukaryotes, extrachromosomal DNA is found in mitochondria and chloroplasts. Mitochondrial DNA replicates via a strand displacement model using DNA polymerase gamma and other specialized replication factors. Chloroplast DNA replication shares similarities but uses its own distinct replication proteins.