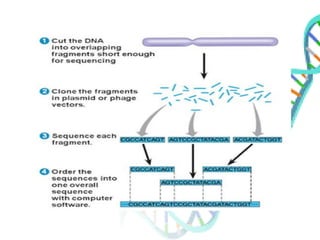
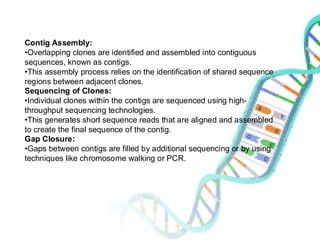
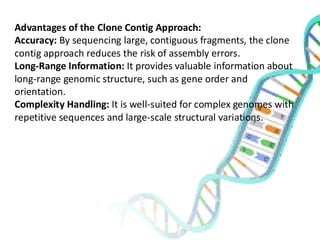
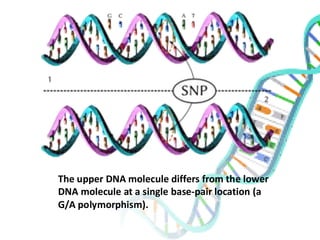
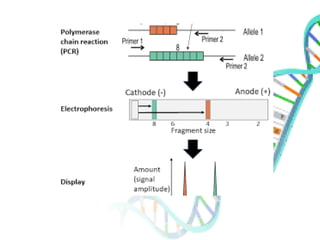
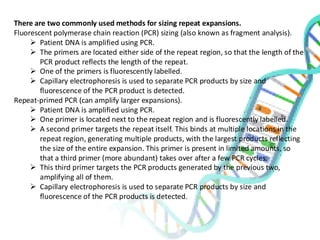
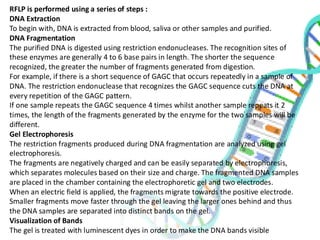
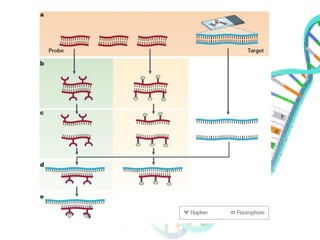
The document provides an extensive overview of genome mapping techniques, including genetic and physical mapping methods such as the shotgun approach, clone contig approach, and various DNA markers like SNPs and STRs. It explains the processes involved in each mapping technique, their advantages, and applications in genetic diagnosis and research. Additionally, it discusses tools such as fluorescence in situ hybridization (FISH) and sequence-tagged sites (STS) mapping, highlighting their specific uses in identifying genetic sequences and disorders.