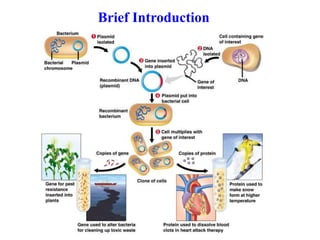
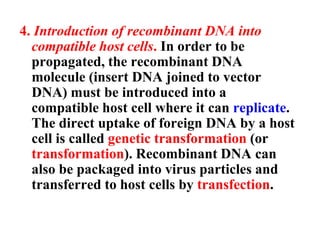
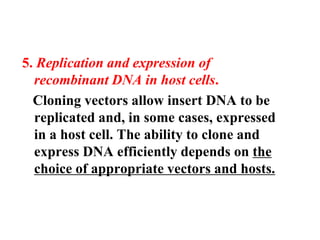
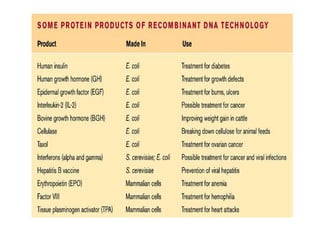
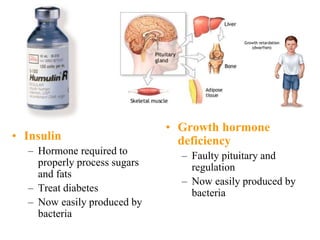
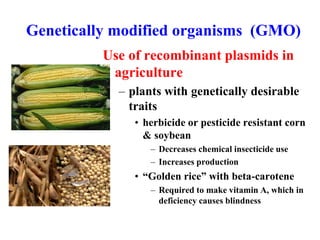
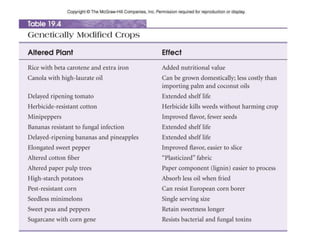
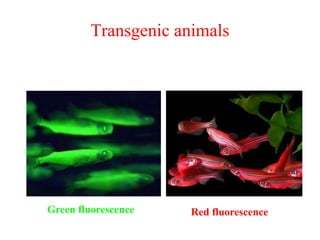
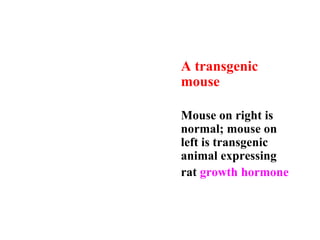
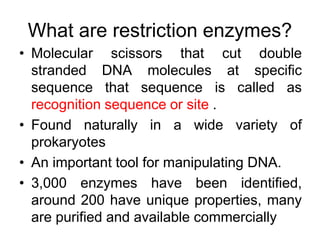
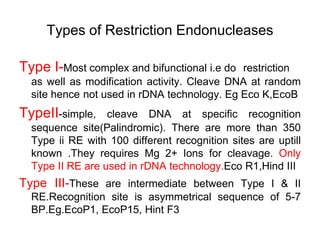
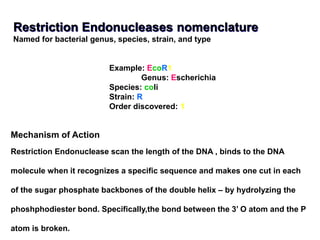
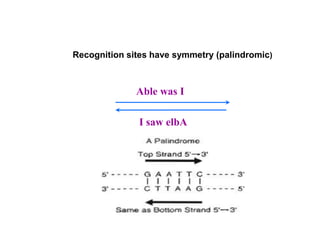
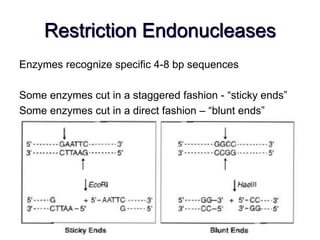
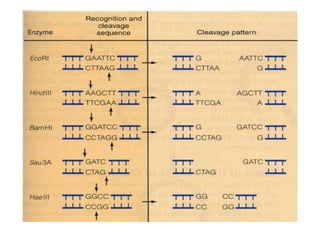
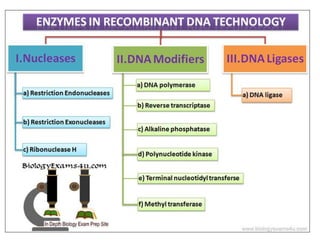
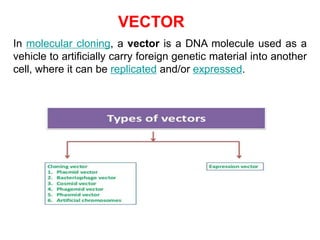
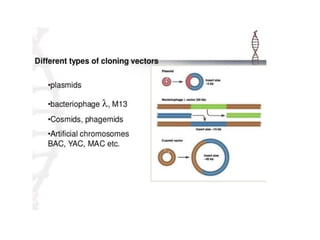
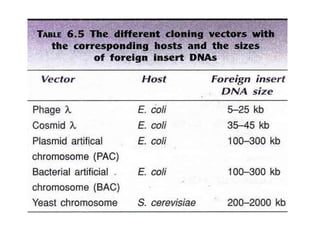
Recombinant DNA technology allows DNA from different sources to be combined to form artificial DNA molecules. This is done by cutting the DNA with restriction enzymes and joining the pieces together with DNA ligase. The artificial DNA can then be inserted into host cells where it is replicated. This technology was developed in 1973 and has many important applications, including producing human insulin in bacteria to treat diabetes, creating genetically modified crops with desirable traits, and producing other proteins and vaccines. The basic steps involve isolating DNA, cutting it with restriction enzymes, ligating the pieces, introducing the DNA into host cells, replicating the DNA within the cells, and identifying cells containing the recombinant DNA.