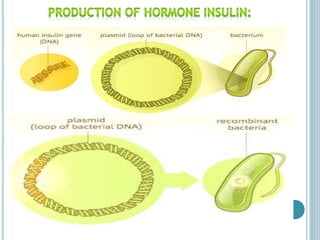
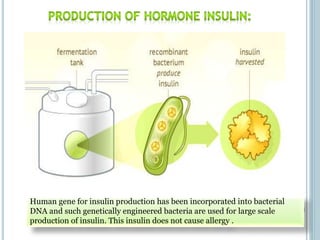
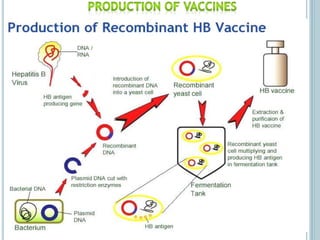
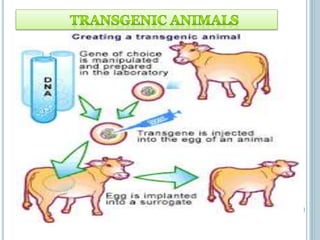
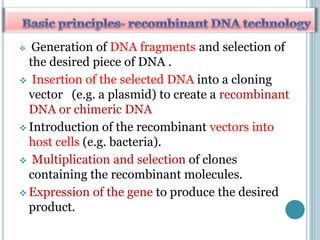
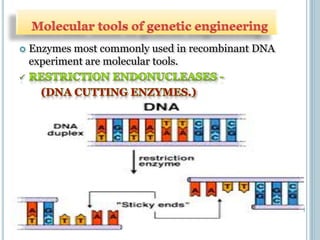
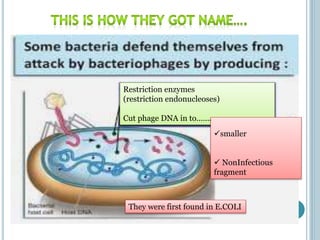
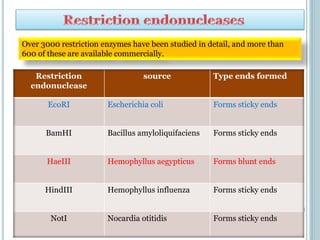
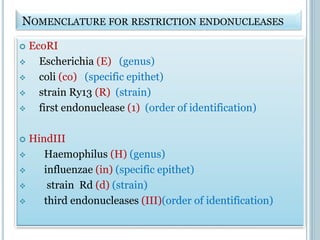
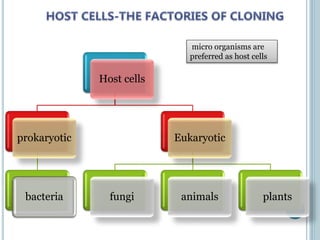
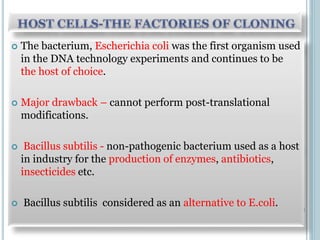
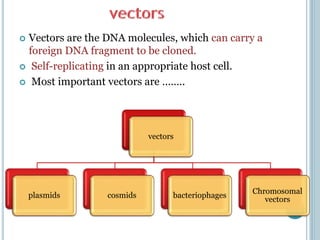
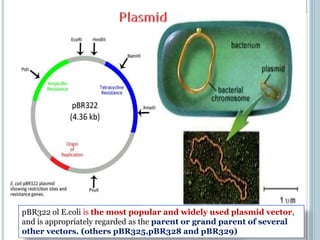
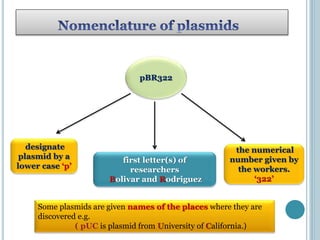
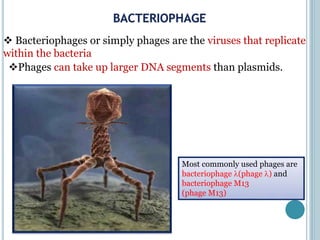
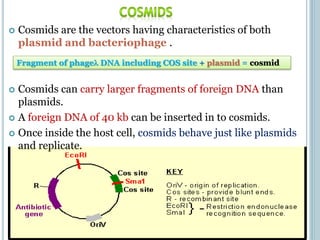
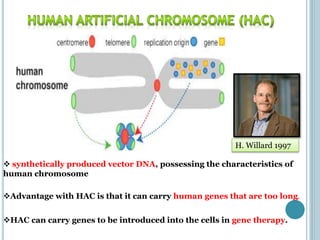
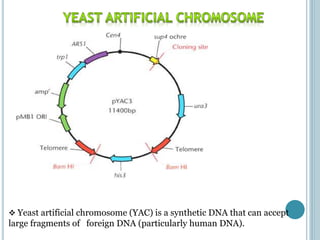
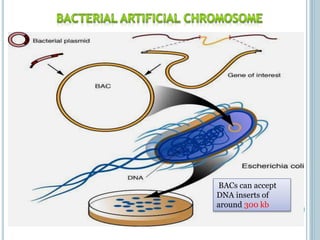
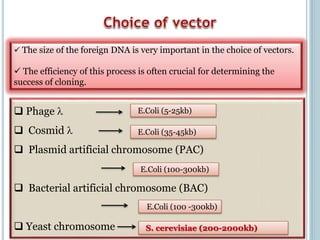
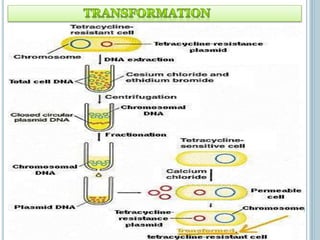
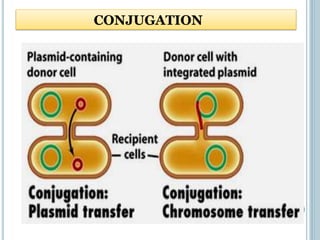
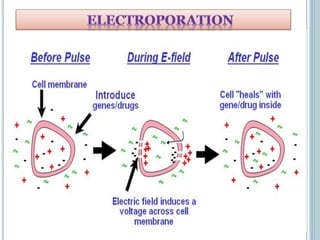
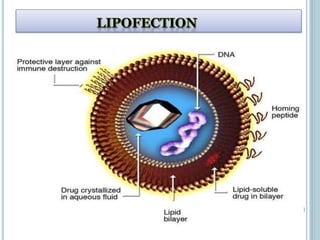
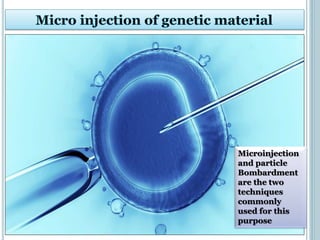
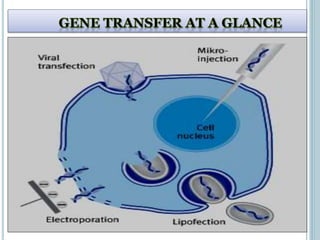
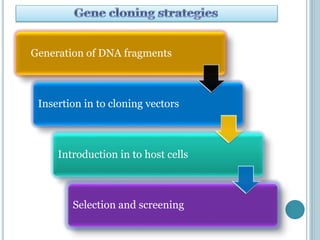
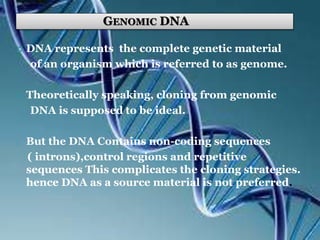
This document discusses recombinant DNA technology and its applications. It summarizes that Herbert Boyer and Stanley Cohen developed recombinant DNA technology in the 1970s, showing that genetically engineered DNA molecules can be cloned in foreign cells. It then provides examples of how recombinant DNA technology is used in agriculture, medicine, and industry for purposes such as producing important proteins and antibiotics, developing disease-resistant crops, and diagnosing diseases.