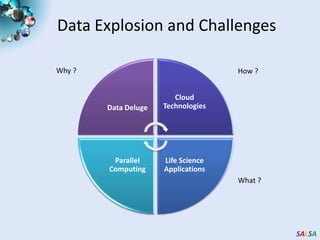
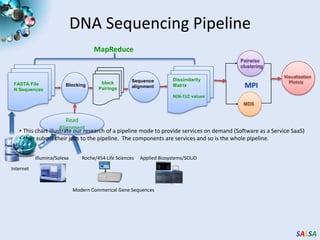
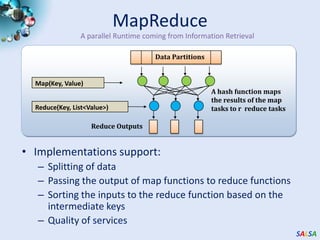
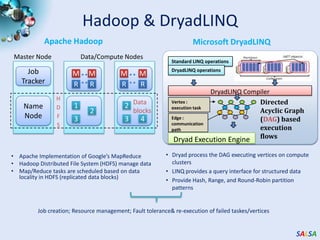
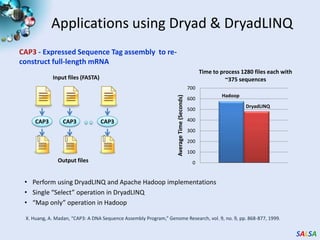
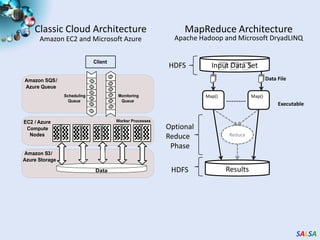
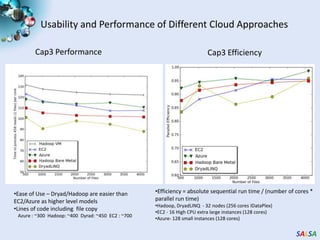
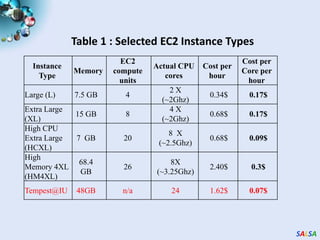
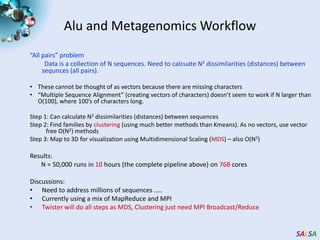
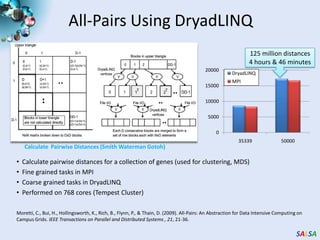
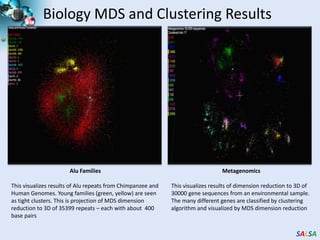
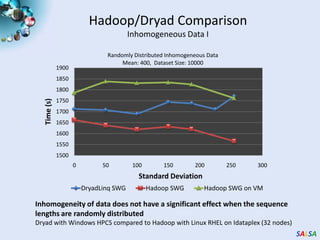
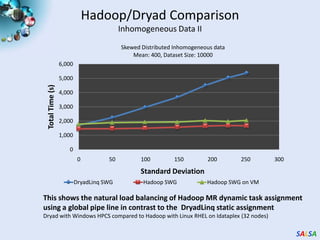
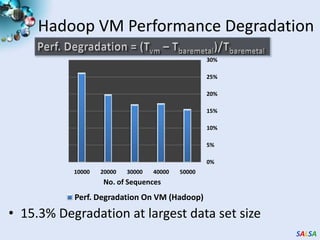
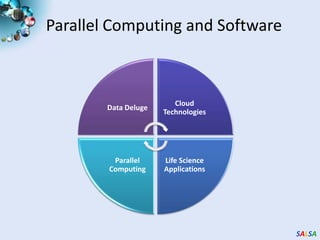
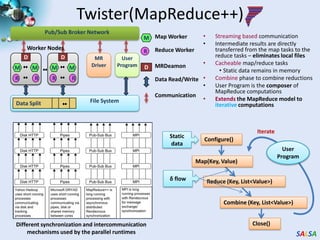
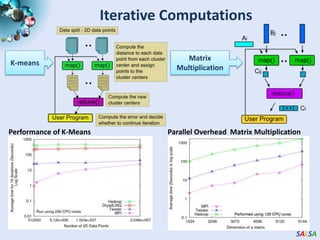
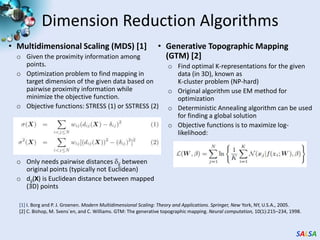
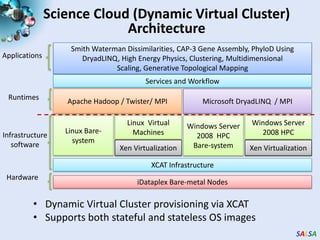
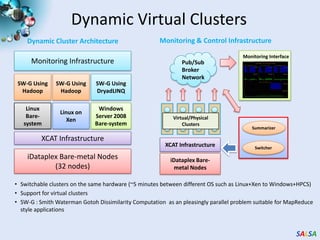
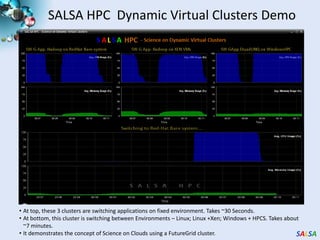
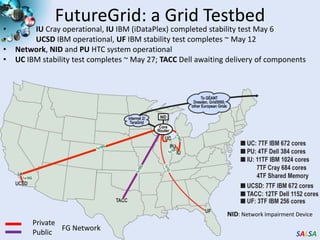
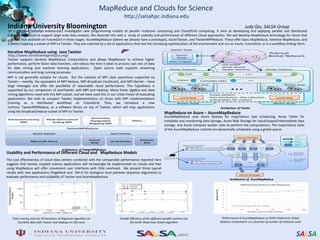
This document summarizes cloud technologies and their applications in life sciences. It discusses how cloud computing can help address challenges posed by big data through cost-effective data centers, hiding complexity, and parallel computing frameworks like MapReduce. Specific applications highlighted include DNA sequence assembly, metagenomics, and correlating health data with environmental factors. Frameworks like Hadoop, DryadLINQ, and Twister are examined for processing large-scale biological data on clouds.