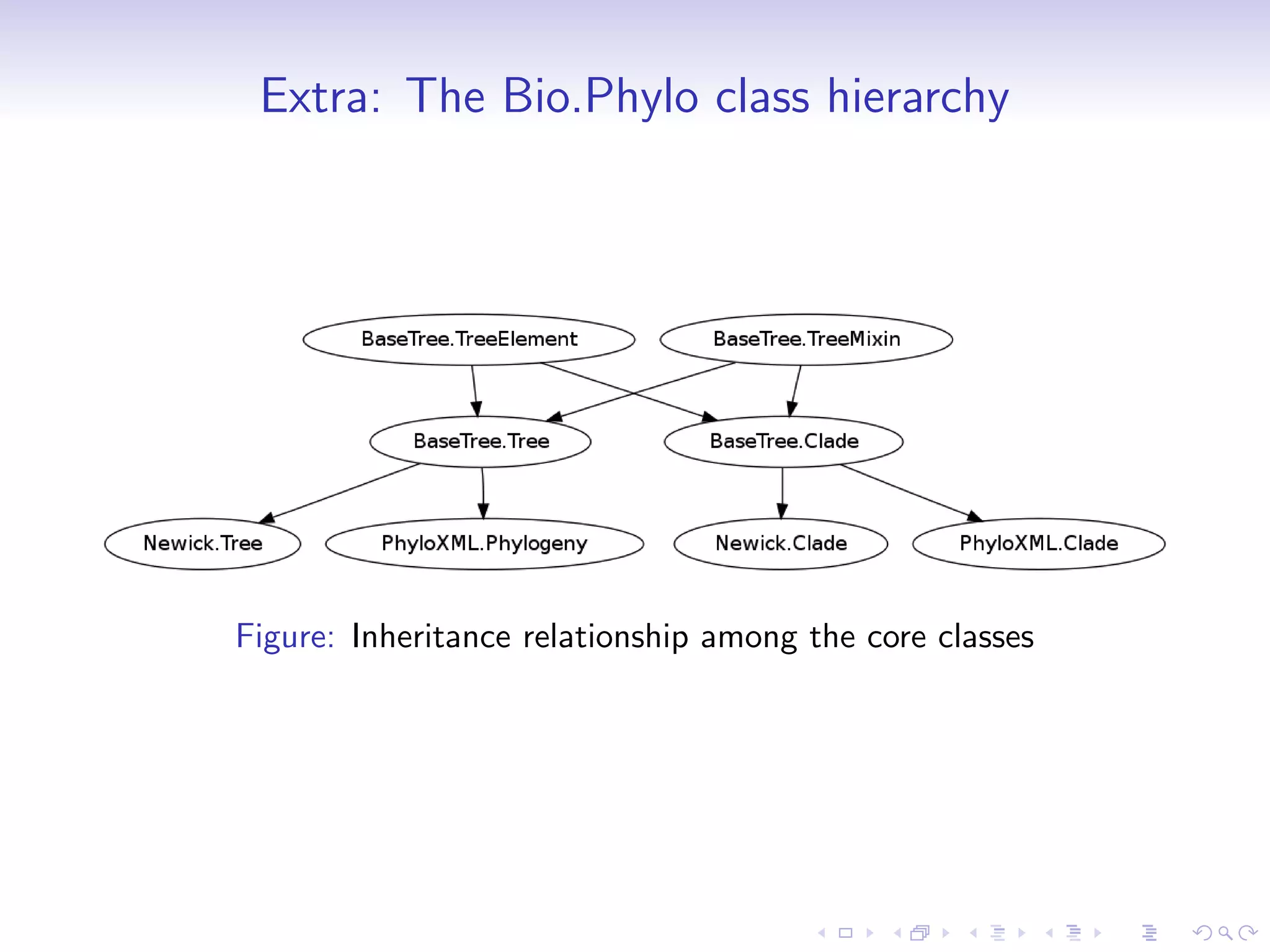
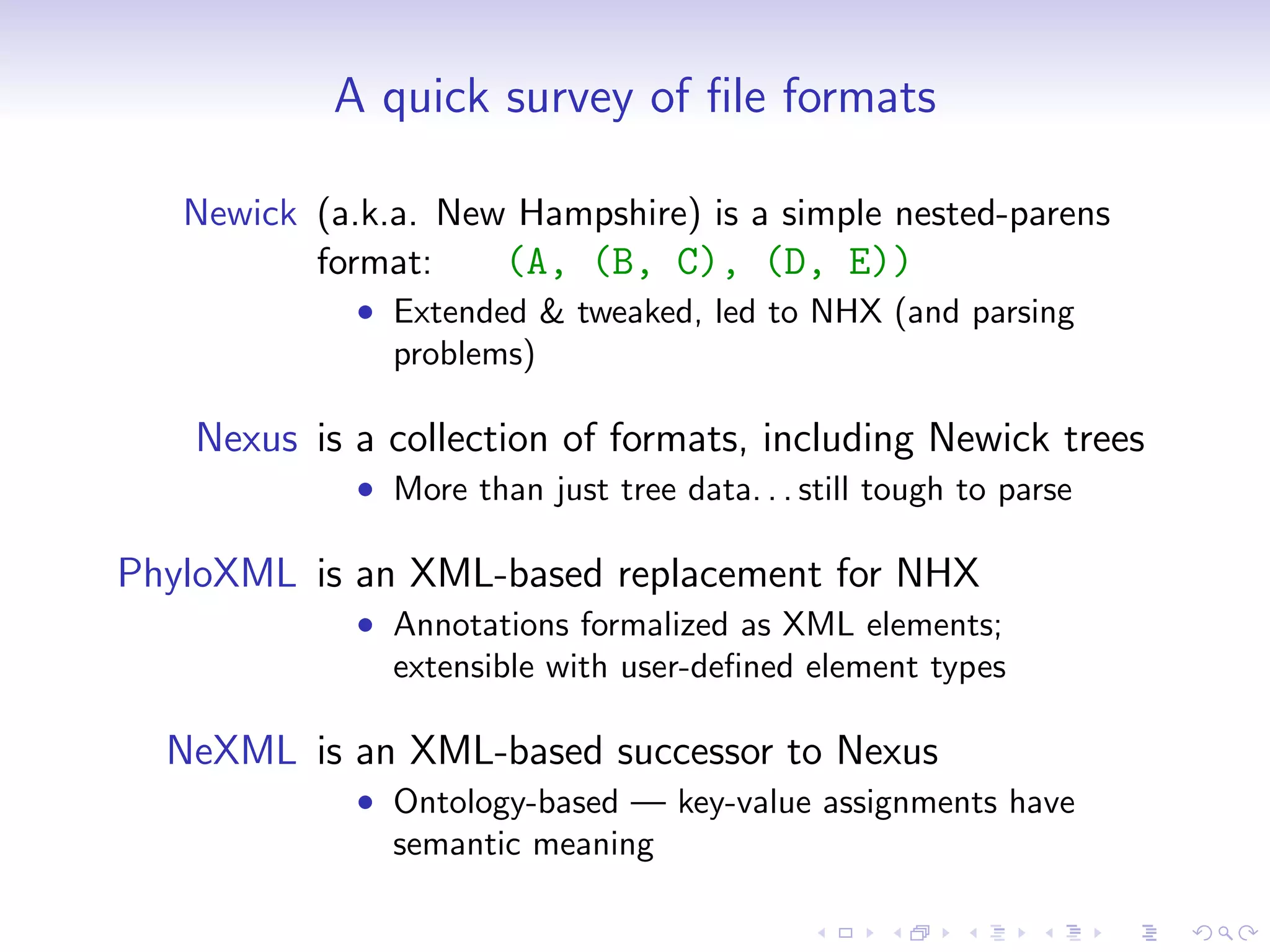
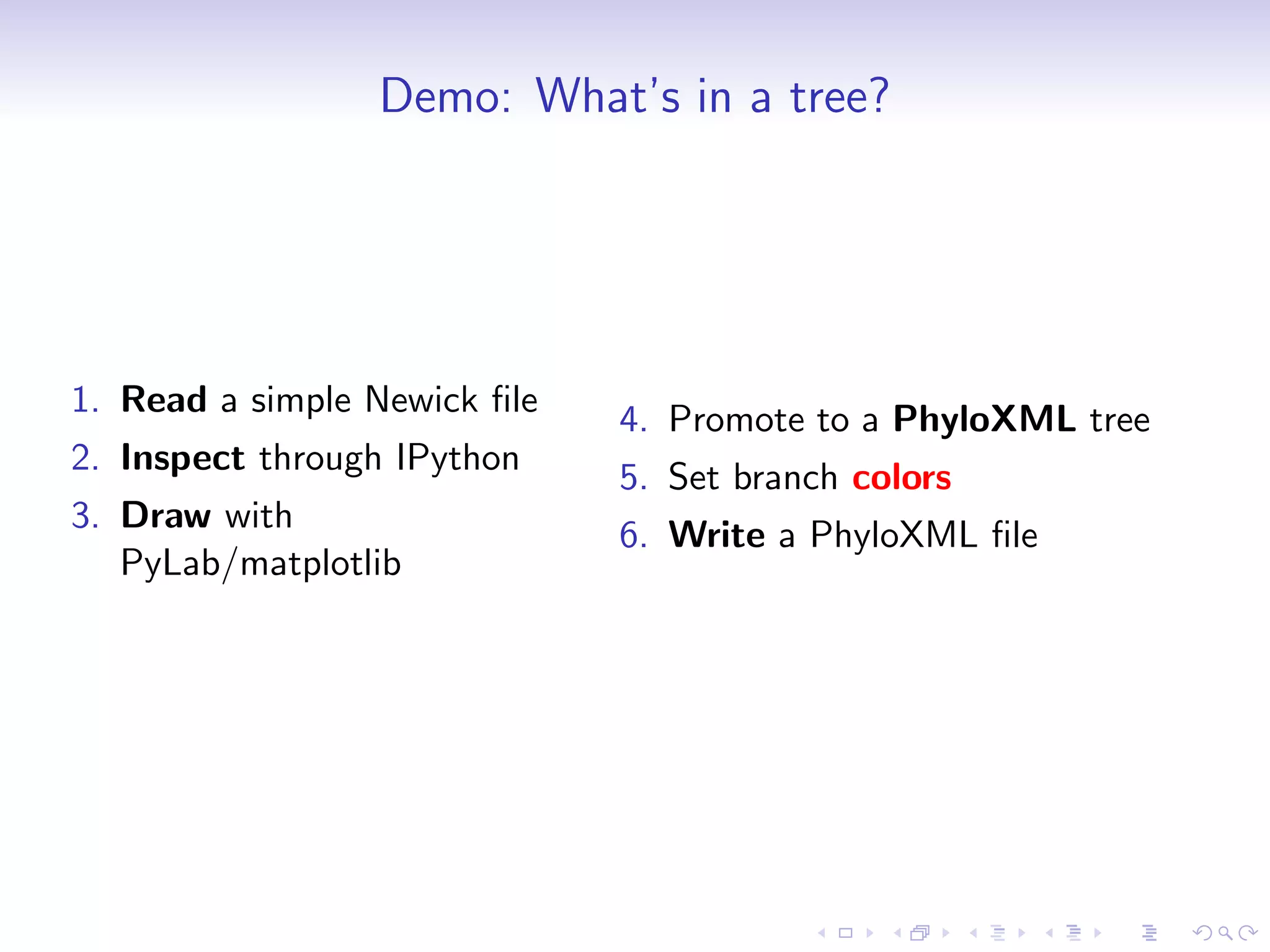
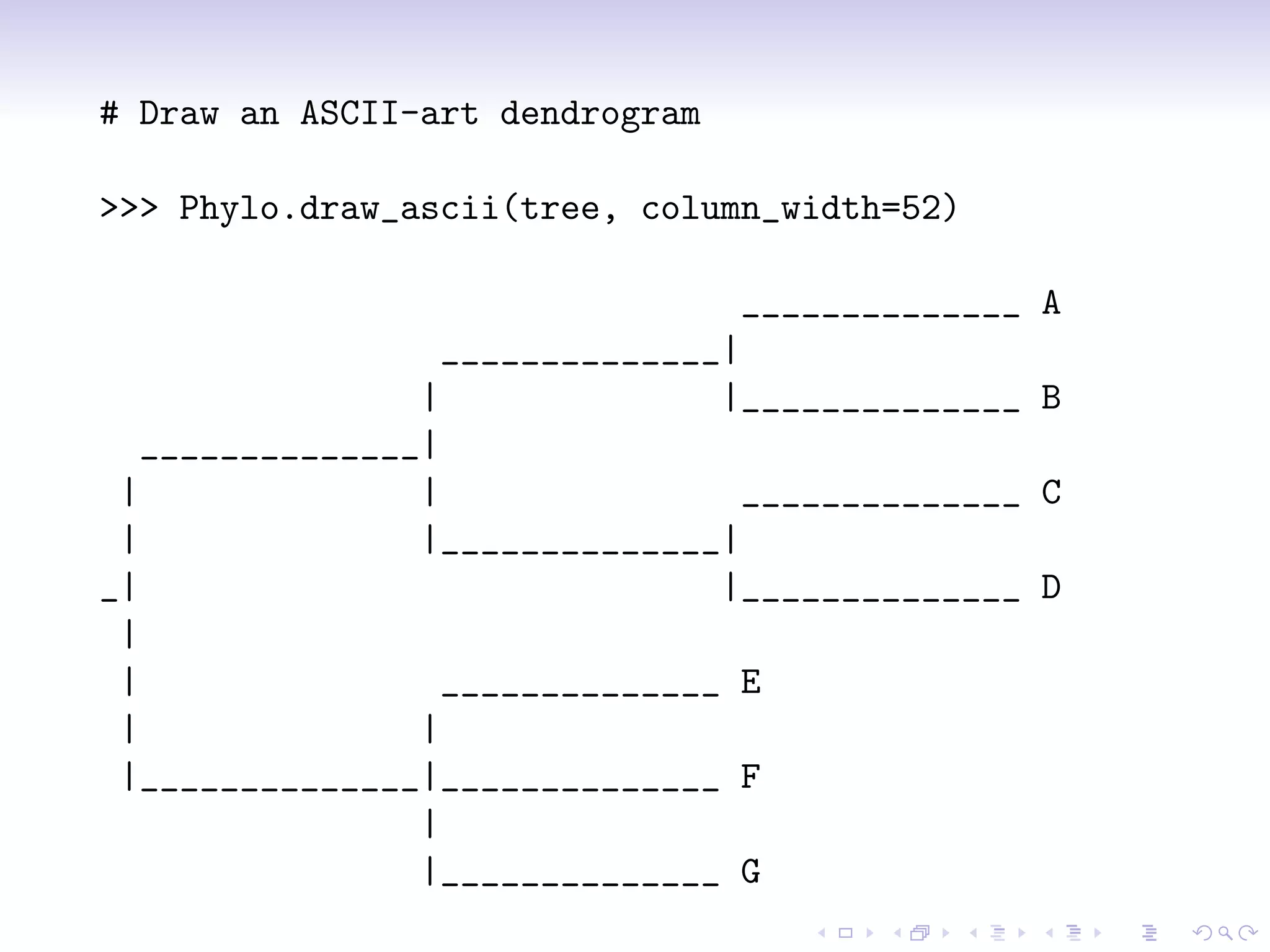
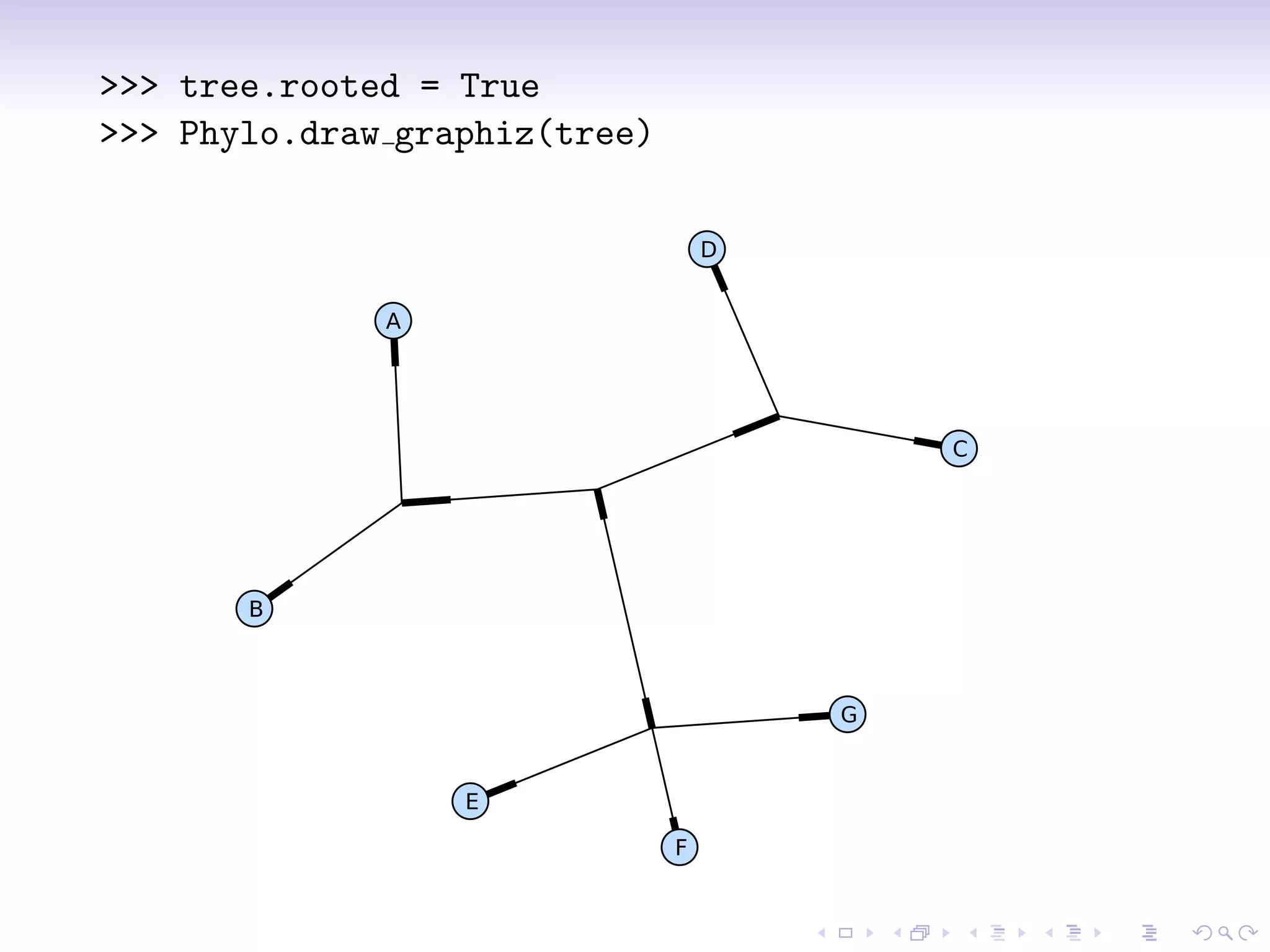
Bio.Phylo is a new phylogenetics library in Biopython for exploring, modifying, annotating, reading, writing, and visualizing trees and for connecting computational pipelines. It supports common file formats like Newick and Nexus and can read/write the XML-based PhyloXML format which allows for annotations. The demo shows how to read a Newick tree, inspect it, draw it, promote it to PhyloXML to add branch colors, and write it out.




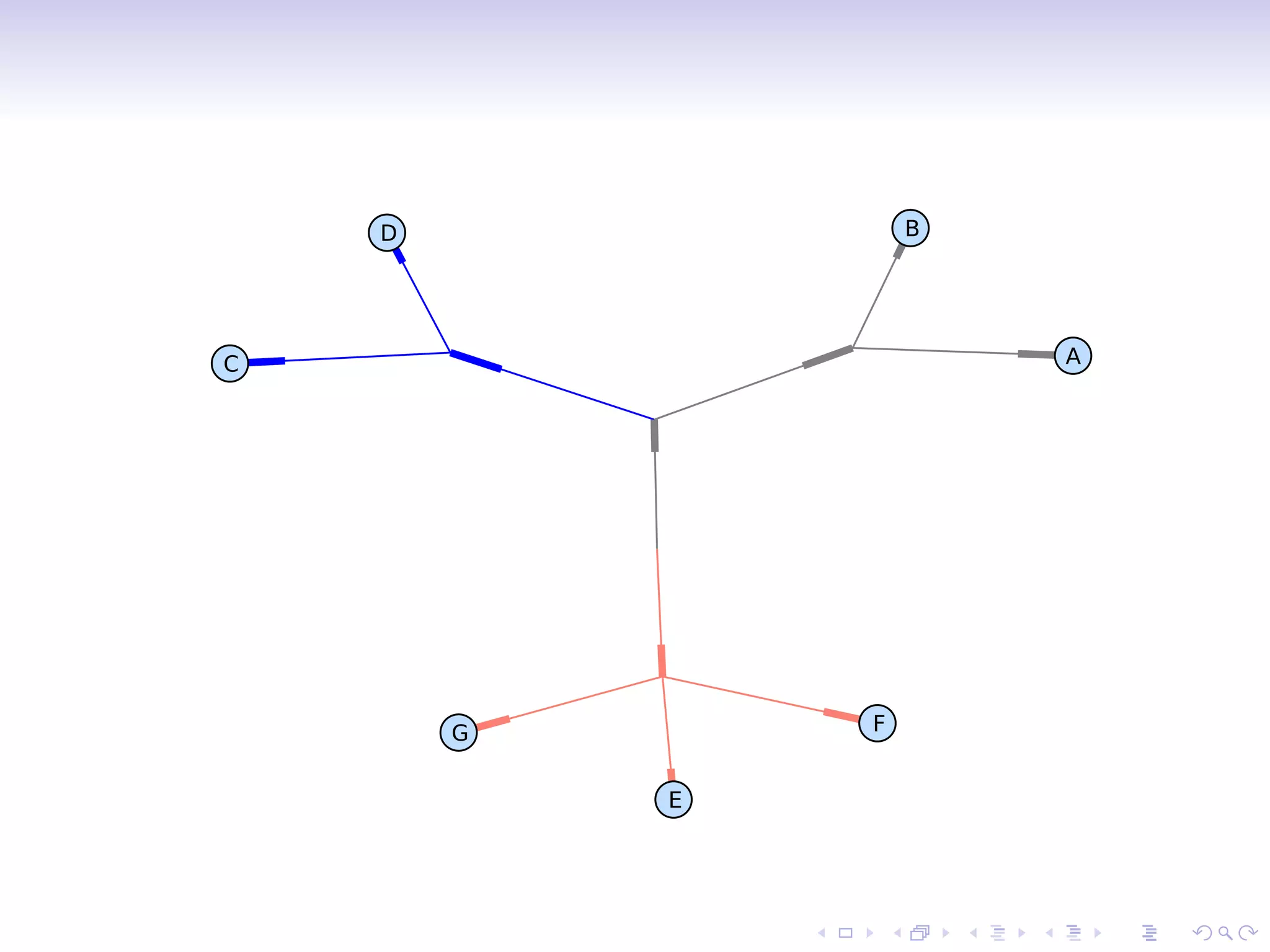
![Branch color
>>> phy.root.color = (128, 128, 128)
Or:
>>> phy.root.color = ’#808080’
Or:
>>> phy.root.color = ’gray’
Find clades by attribute values:
>>> mrca = phy.common ancestor({’name’:’E’},
{’name’:’F’})
>>> mrca.color = ’salmon’
Directly index a clade:
>>> phy.clade[0,1].color = ’blue’
>>> Phylo.draw graphviz(phy, prog=’neato’)](https://image.slidesharecdn.com/talevichbosc2010-biophylo-100710215828-phpapp01/75/Talevich-bosc2010-bio-phylo-10-2048.jpg)