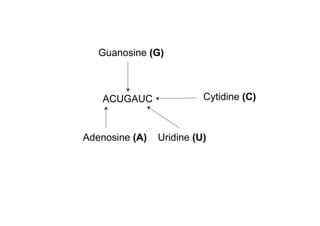
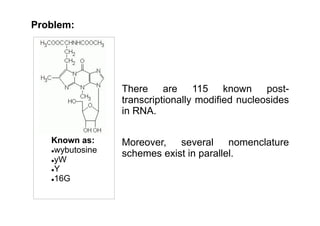
This document discusses BioPython modules for handling RNA sequences containing modified nucleosides. There are 115 known post-transcriptionally modified nucleosides in RNA and several nomenclature schemes exist. The solution involves cloning a branch of the BioPython repository containing an RNA alphabet with modified nucleotides and using it to represent sequences containing modifications like 2-O-methyloadenosine. Example applications presented are ModeRNA for RNA structure modeling and CompaRNA for benchmarking RNA structure prediction methods, both of which use open source tools including BioPython.




![Solution:
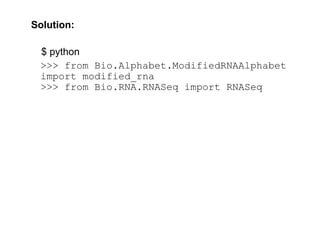
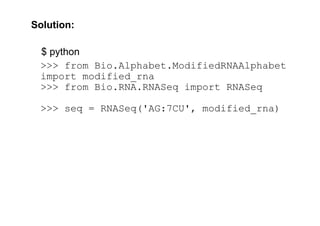
$ python
>>> from Bio.Alphabet.ModifiedRNAAlphabet
import modified_rna
>>> from Bio.RNA.RNASeq import RNASeq
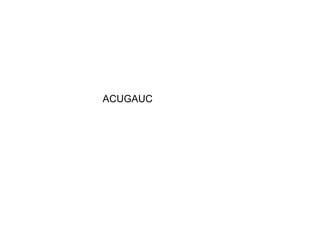
>>> seq = RNASeq('AG:7CU', modified_rna)
>>> print seq[2].full_name
2-O-methyloadenosine](https://image.slidesharecdn.com/putonbosc2010biopython-modules-rna-100710215850-phpapp01/85/Puton-bosc2010-bio_python-modules-rna-11-320.jpg)
![Solution:
$ python
>>> from Bio.Alphabet.ModifiedRNAAlphabet
import modified_rna
>>> from Bio.RNA.RNASeq import RNASeq
>>> seq = RNASeq('AG:7CU', modified_rna)
>>> print seq[2].full_name
2-O-methyloadenosine
>>> print seq[3].long_abbrev
m7G](https://image.slidesharecdn.com/putonbosc2010biopython-modules-rna-100710215850-phpapp01/85/Puton-bosc2010-bio_python-modules-rna-12-320.jpg)