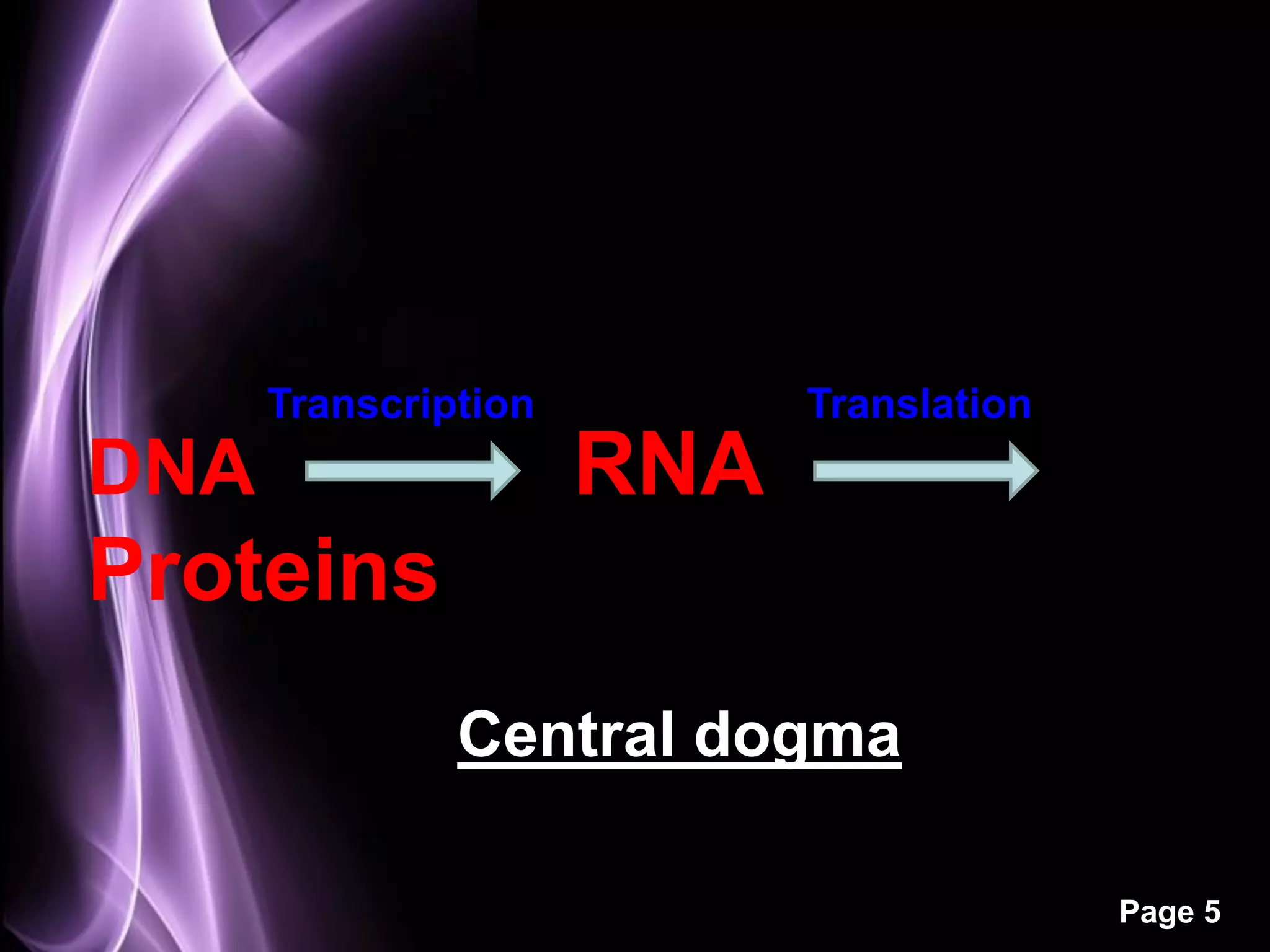
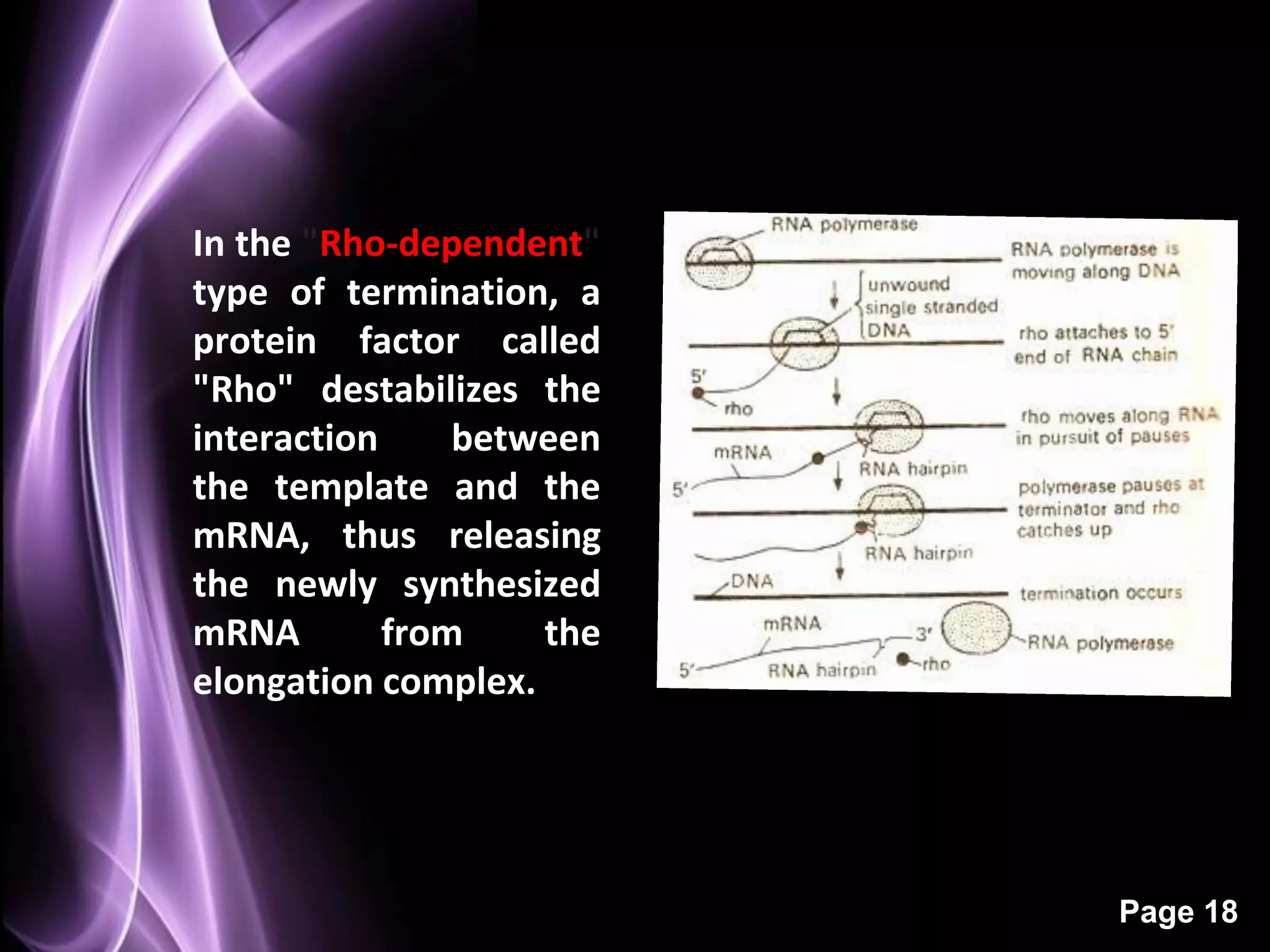
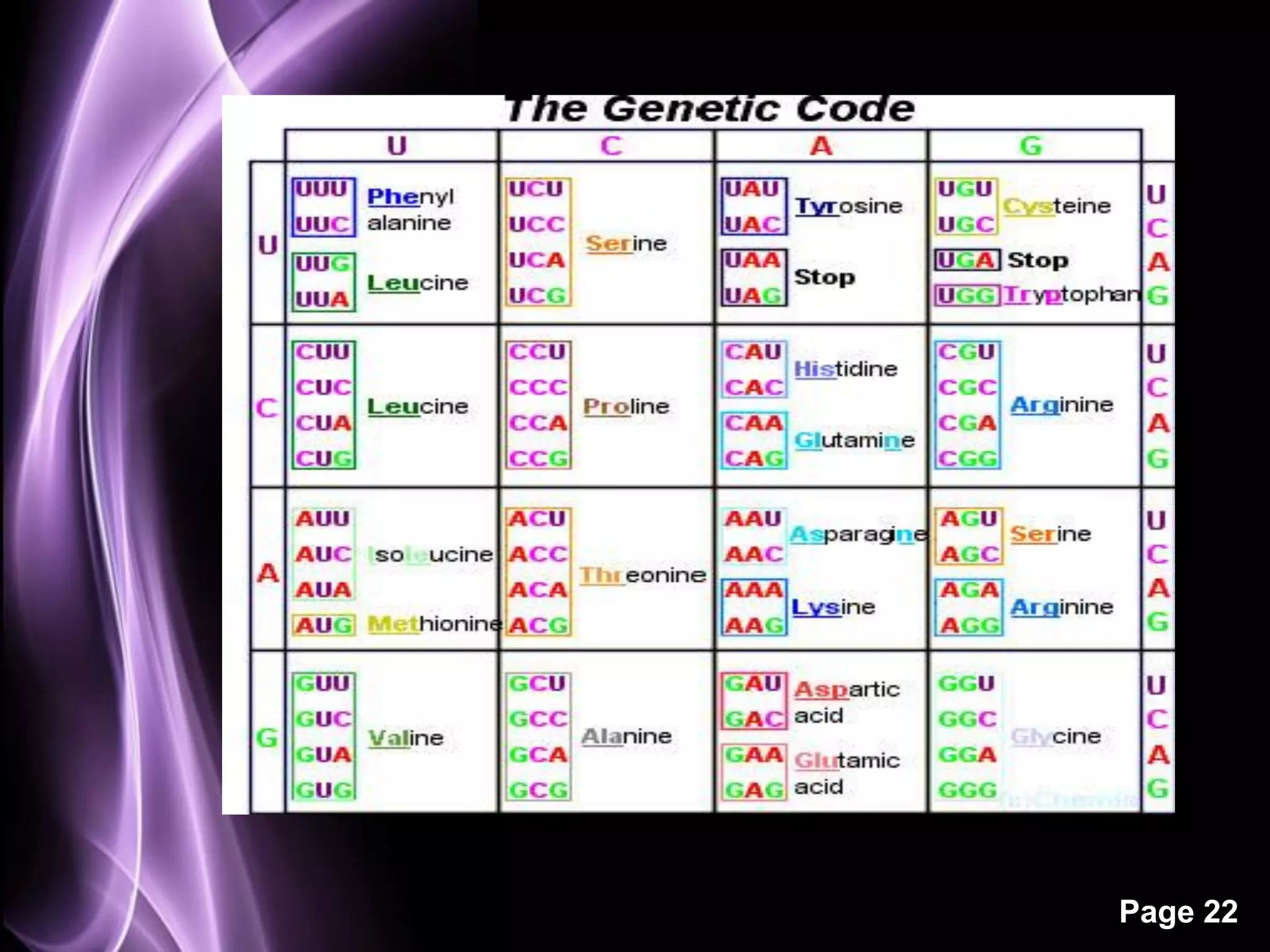
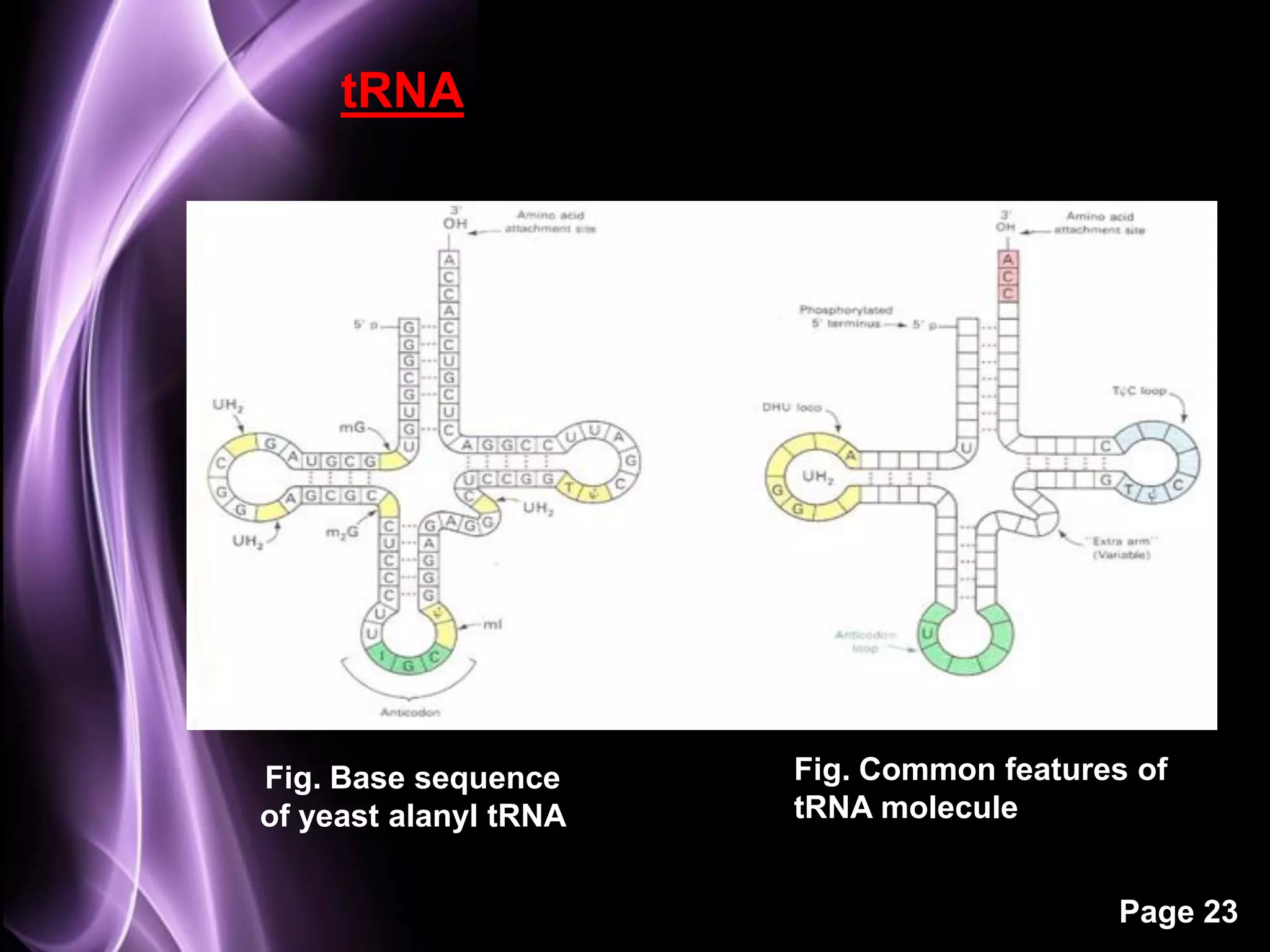
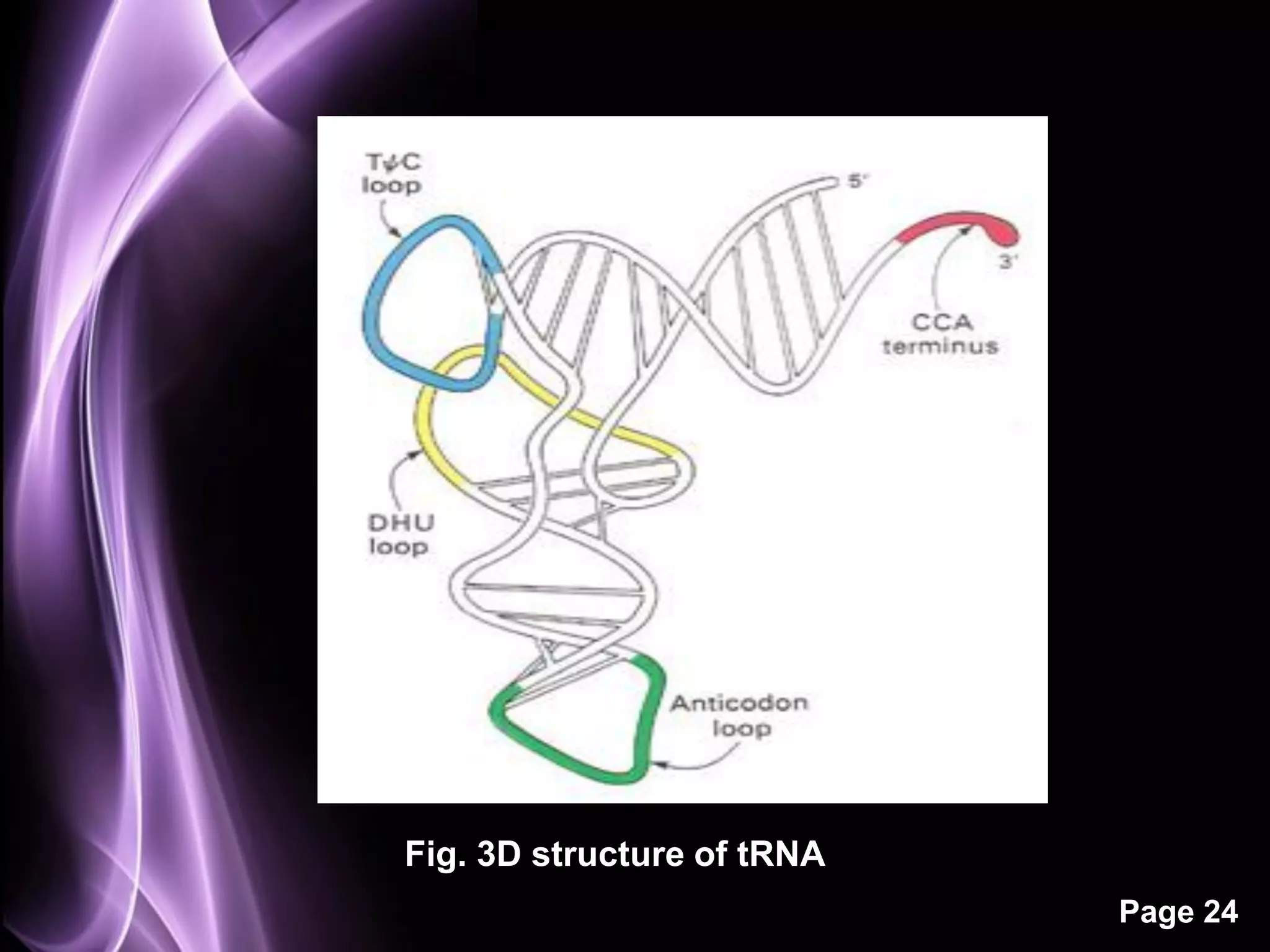
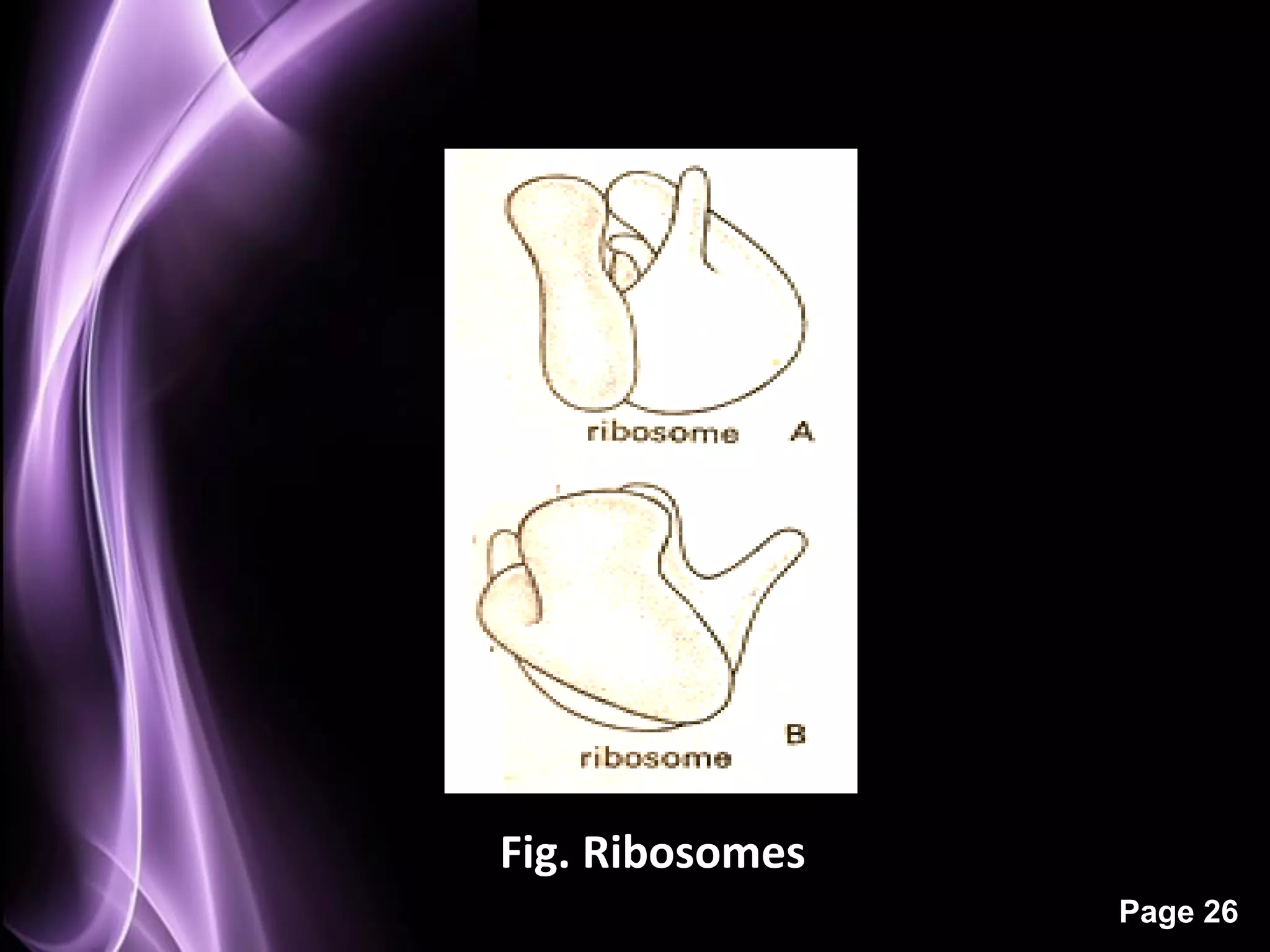
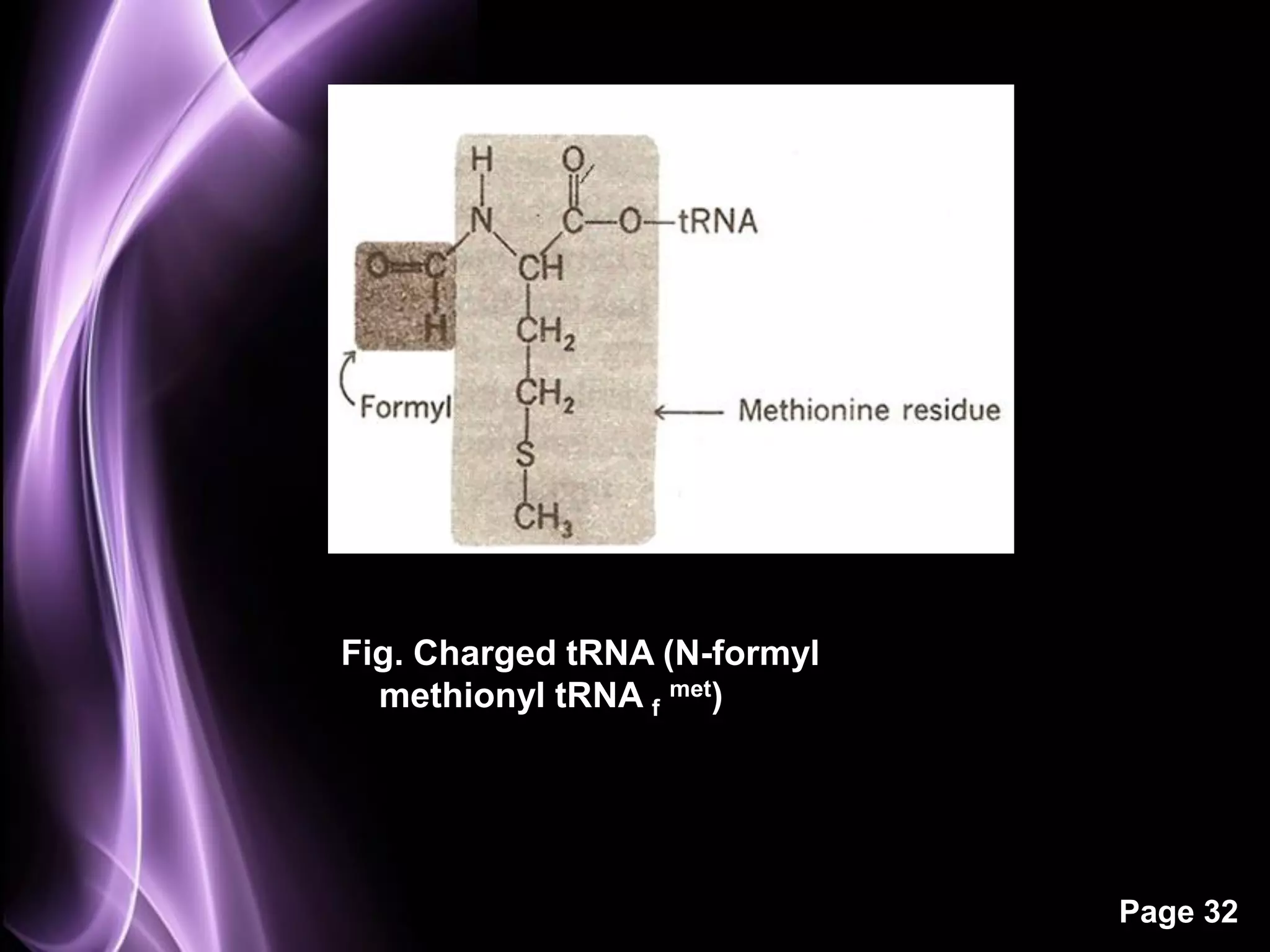
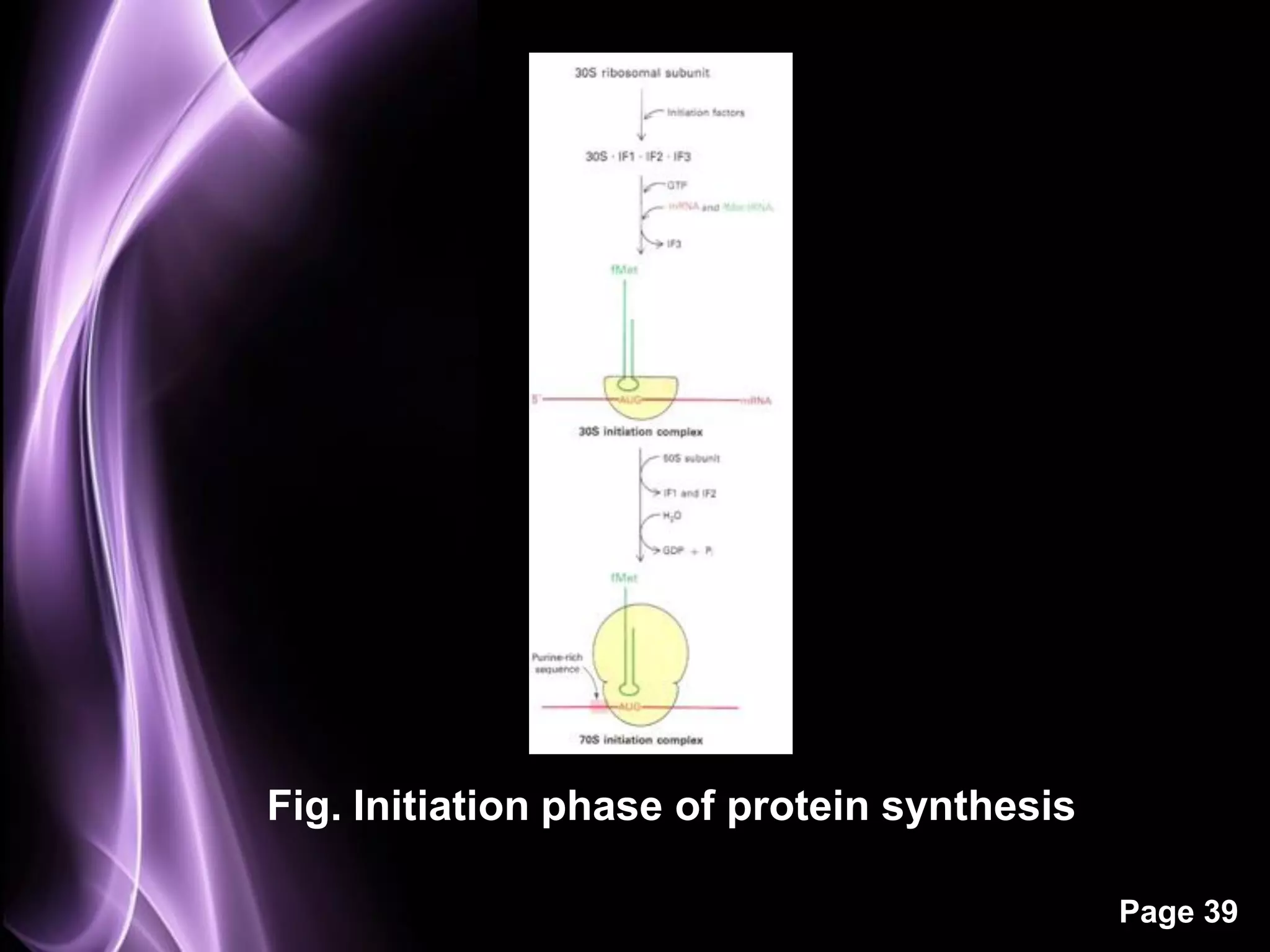
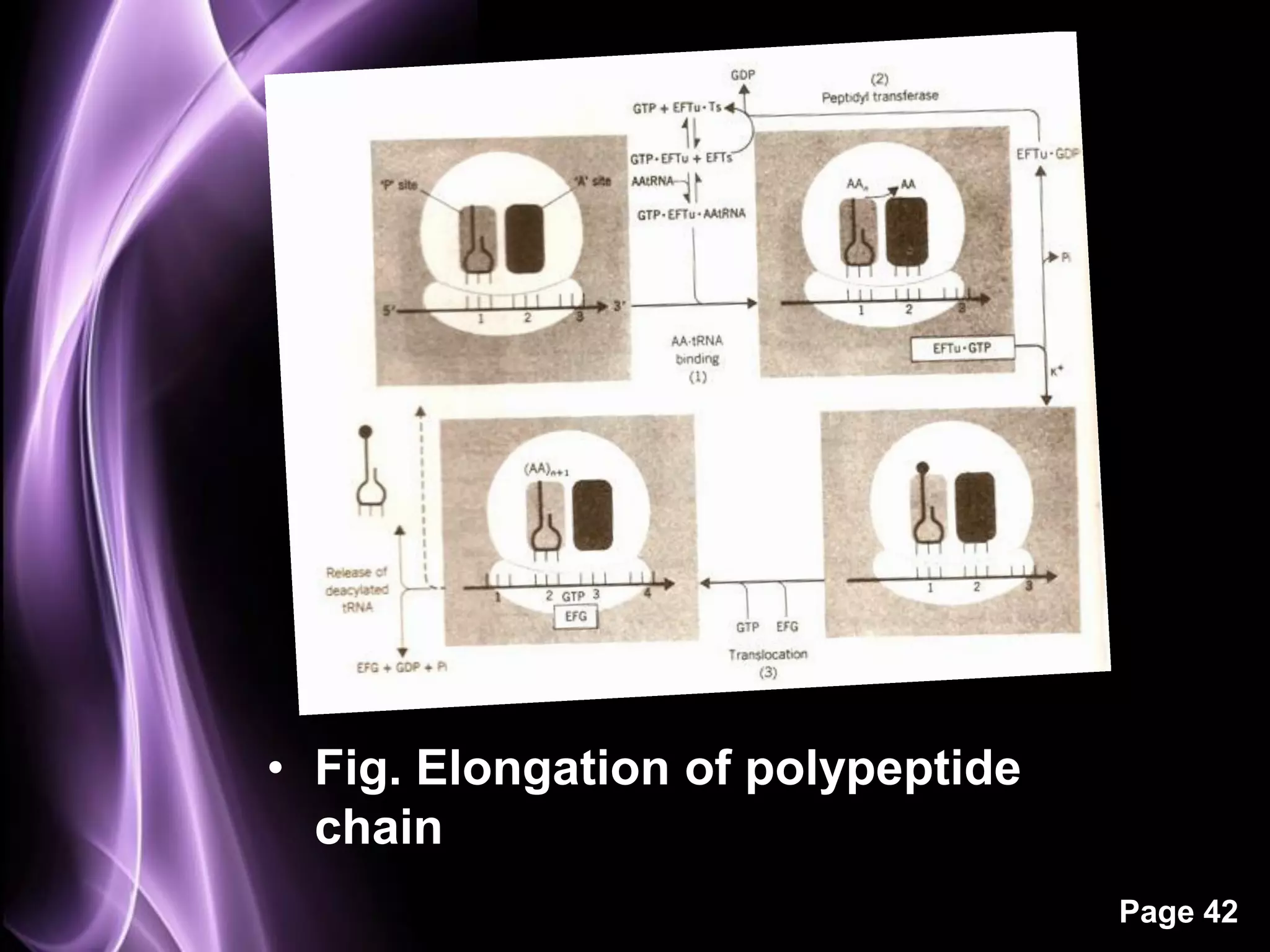
The document summarizes the process of protein synthesis in three main steps: transcription, translation, and termination. During transcription, RNA polymerase makes an mRNA copy of a DNA sequence. Translation then uses the mRNA to assemble a polypeptide chain via tRNAs and ribosomes. Termination occurs when a stop codon signals the release of the completed protein chain. The central dogma of biology is demonstrated as DNA is transcribed to mRNA which is then translated to protein.