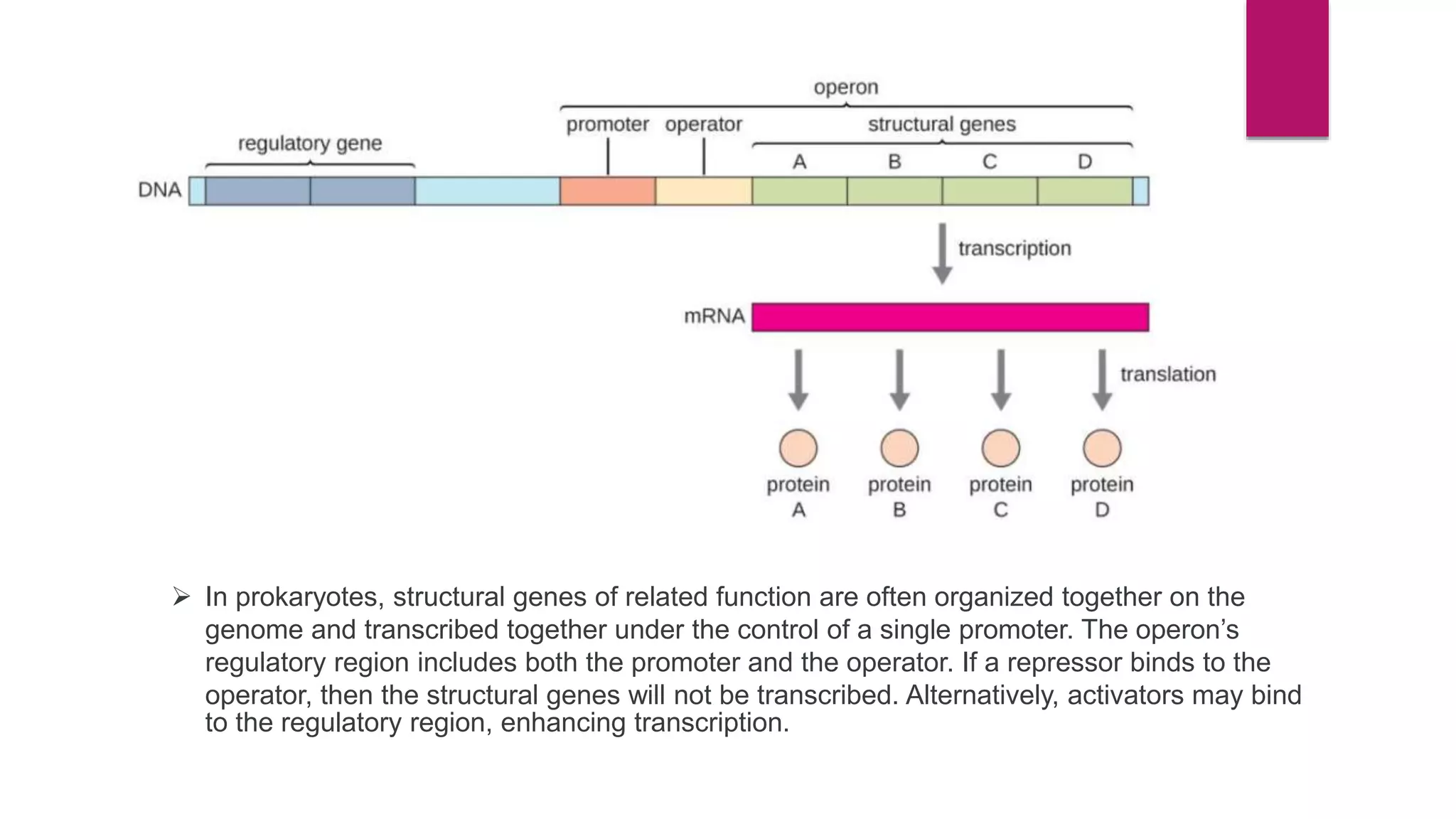
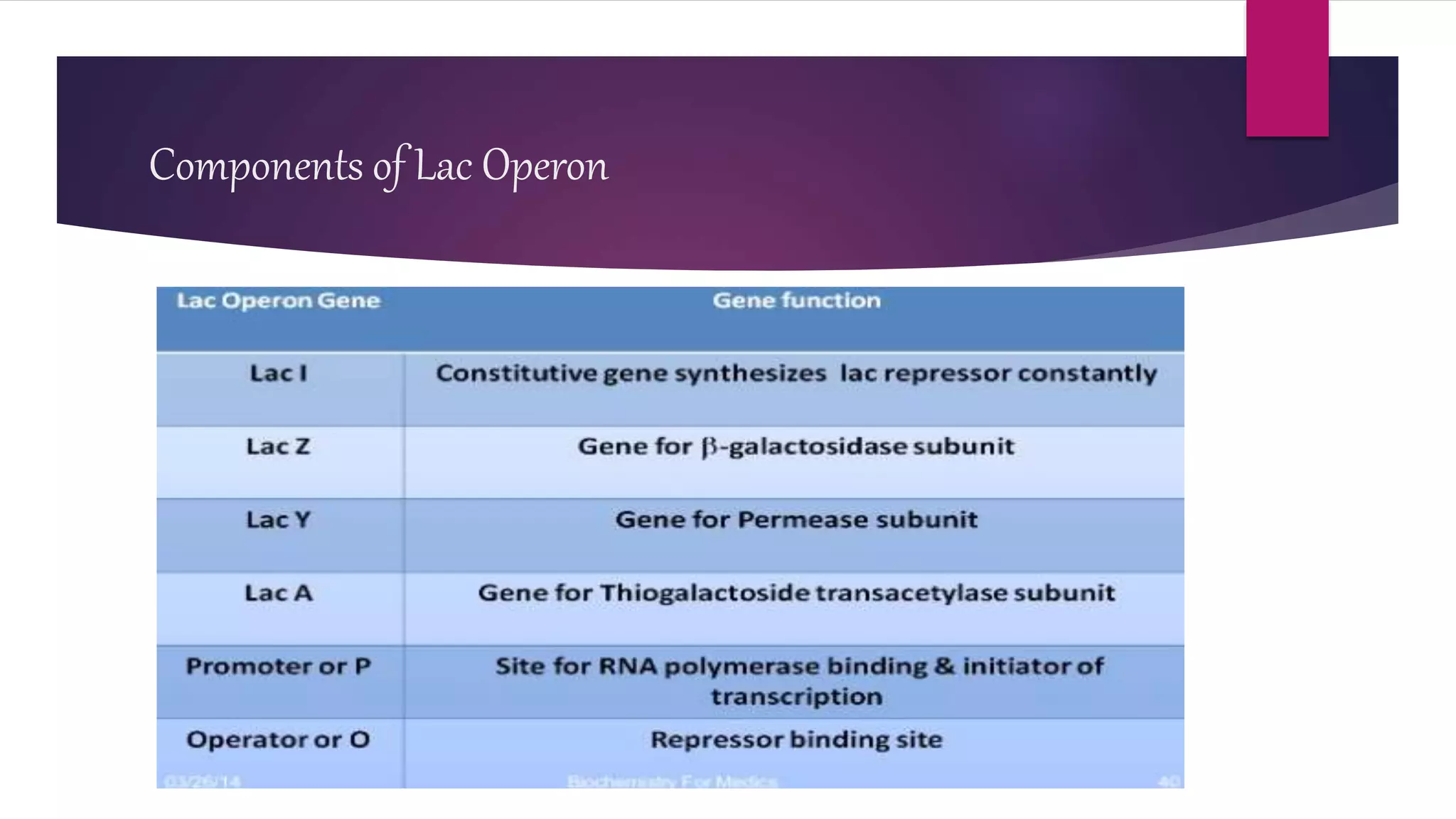
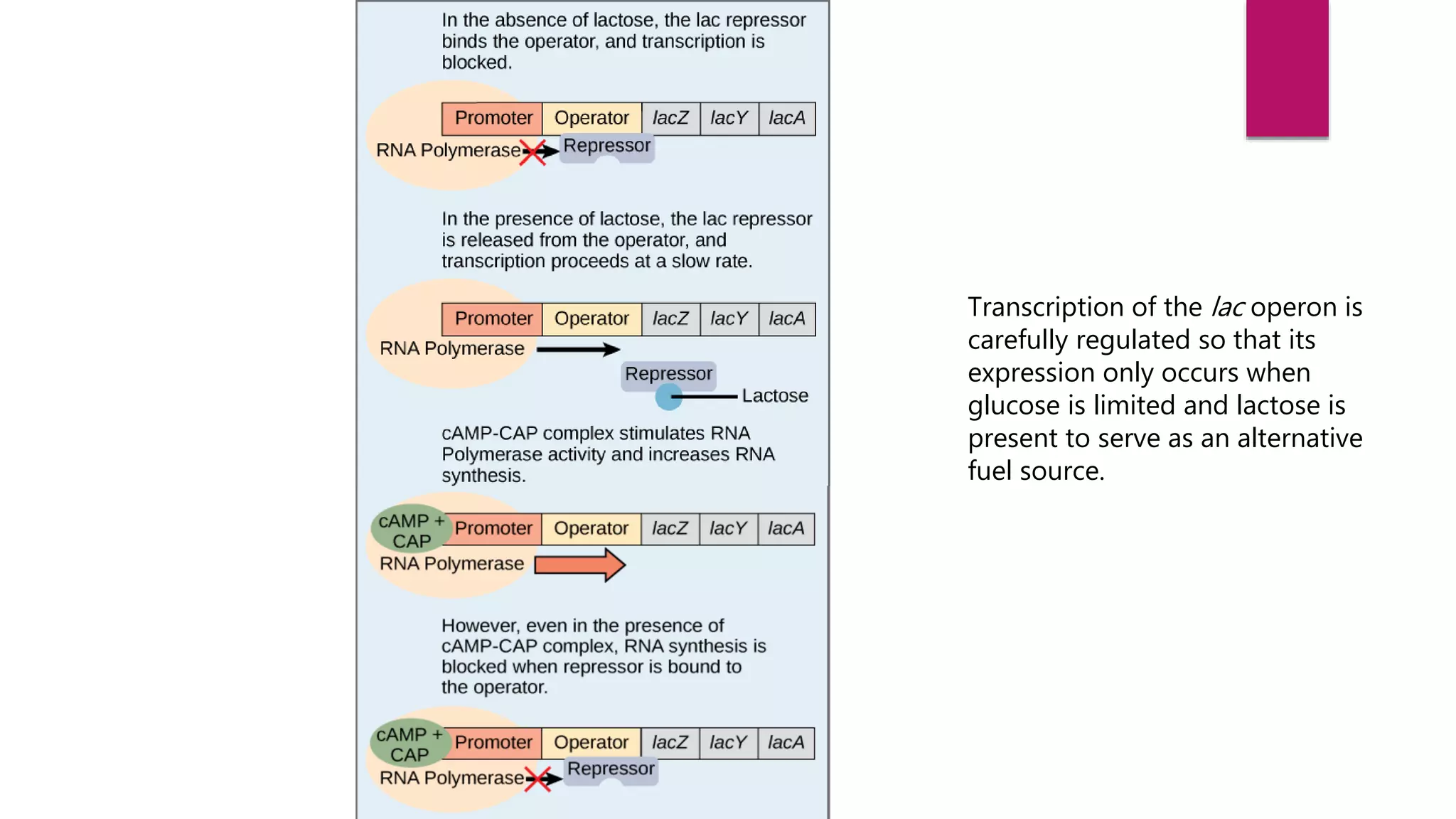
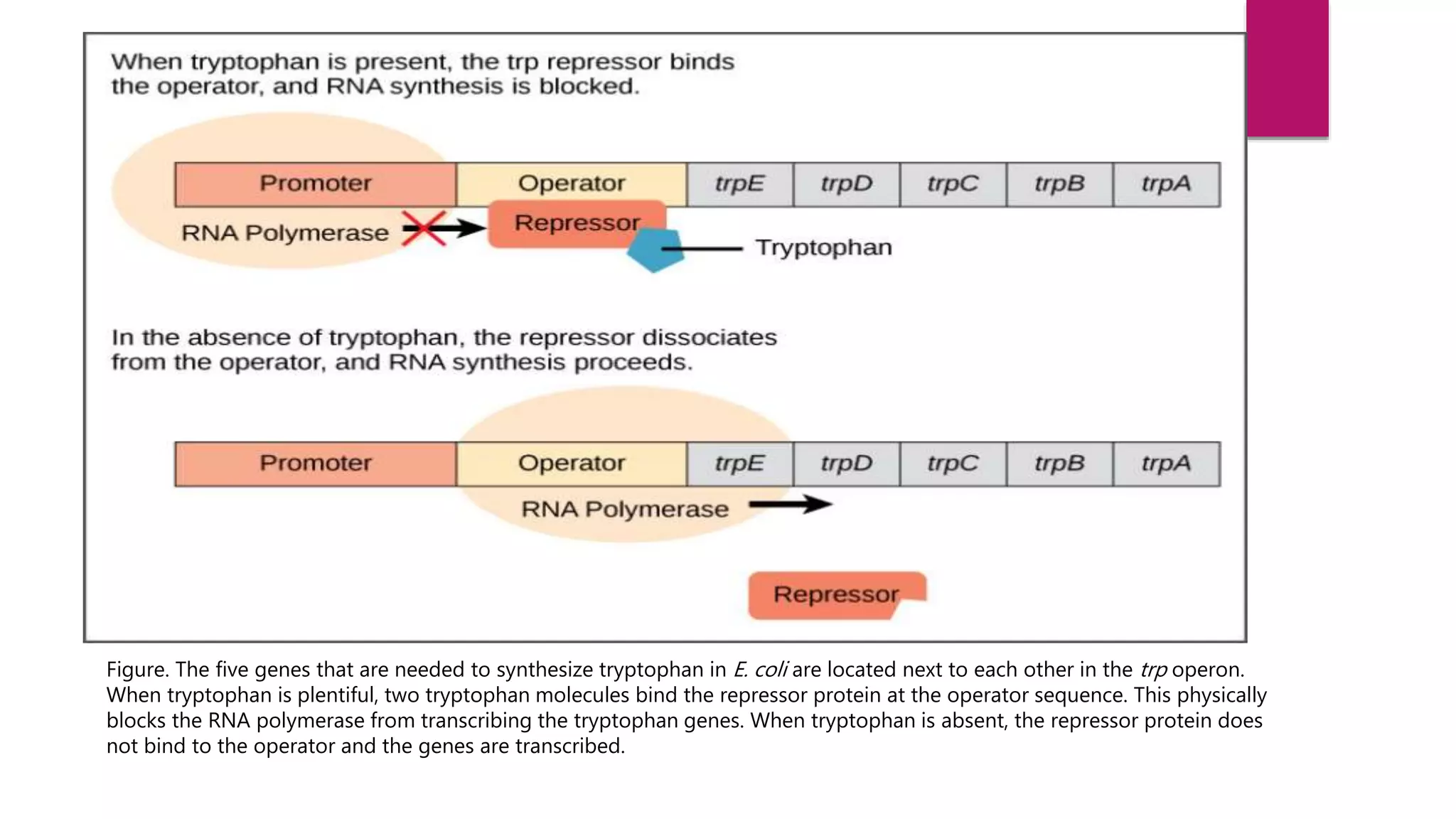
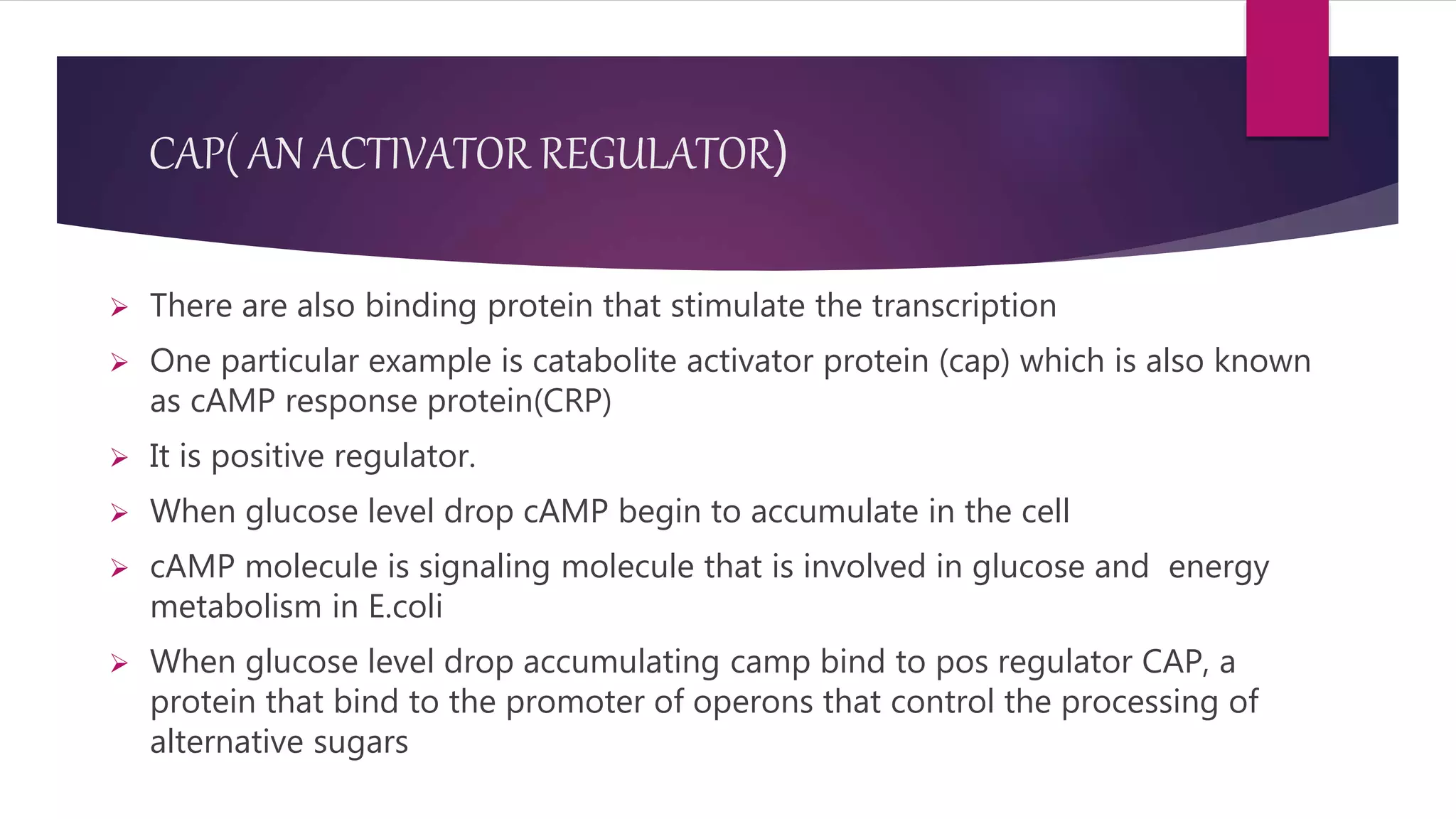
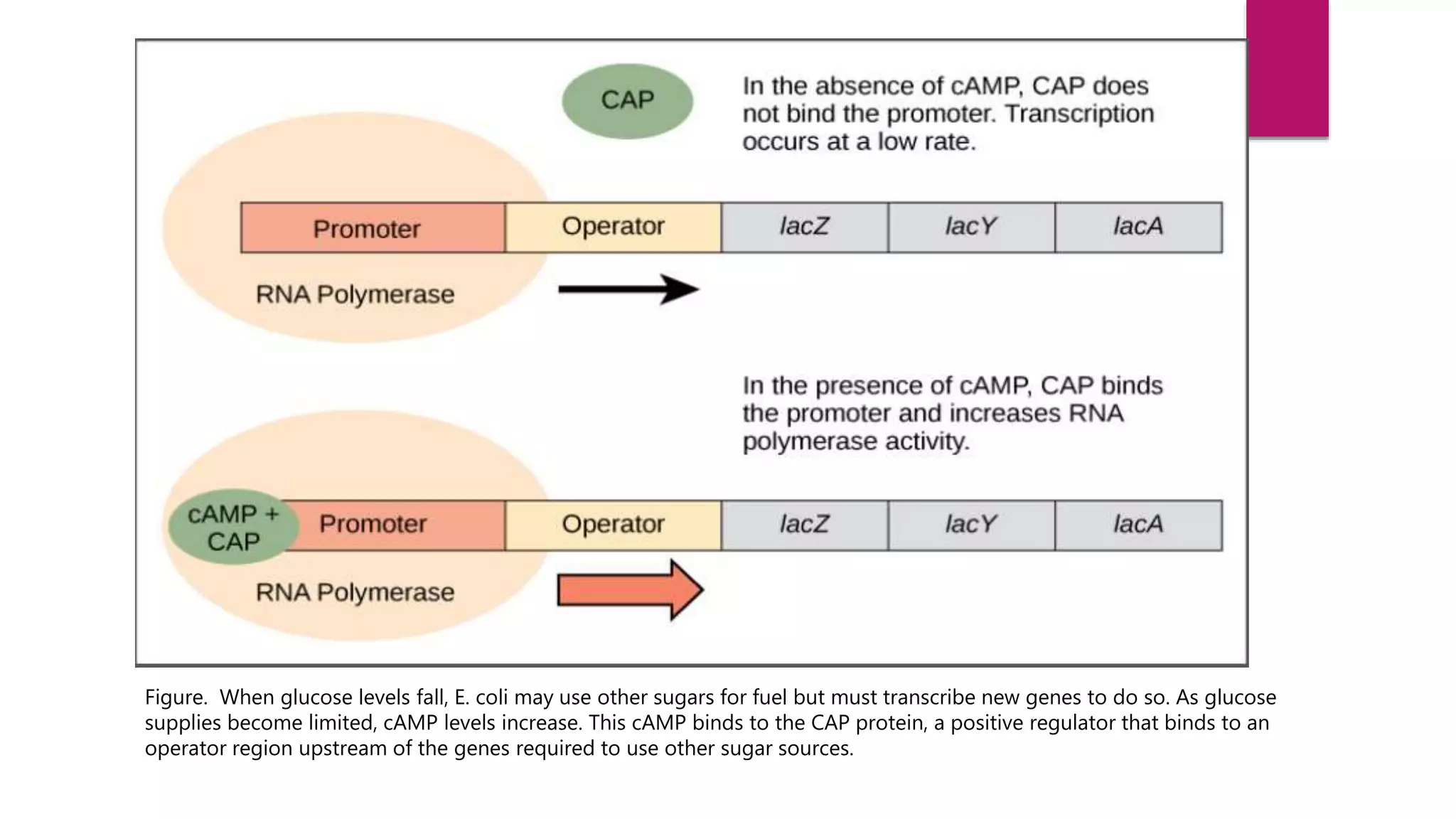
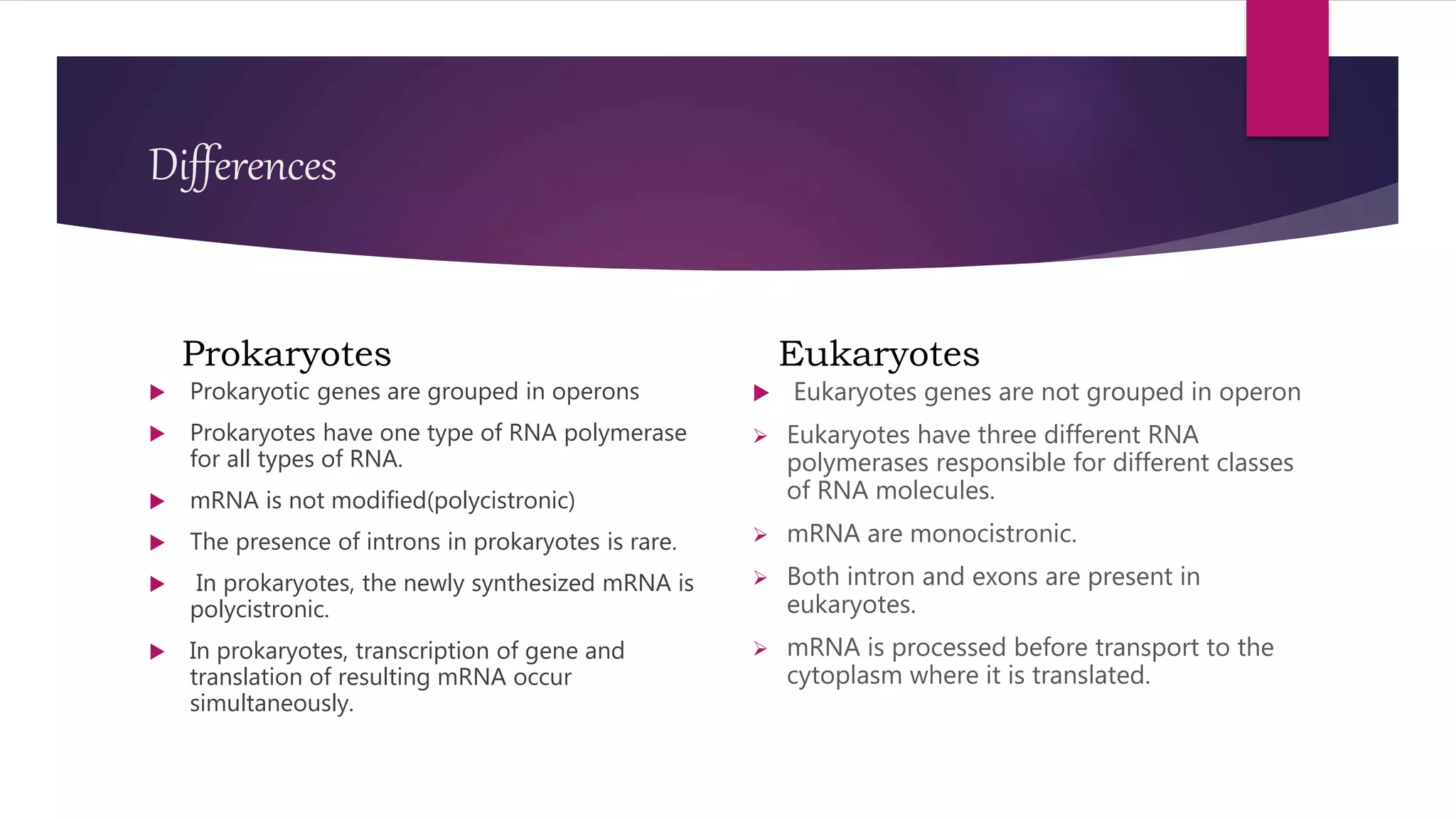
The document discusses prokaryotic expression systems, which are genetic constructs used to produce proteins or RNA inside or outside cells. It focuses on the prokaryotic system, which is most widely used for recombinant protein production in E. coli bacteria. Key aspects covered include the history of prokaryotic expression research, types of gene regulation like operons and regulatory molecules, examples of operons like the lac and trp operons, and differences between prokaryotic and eukaryotic gene expression.