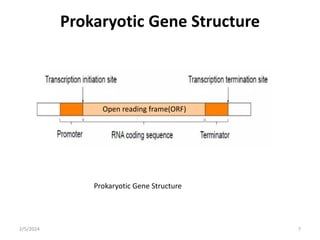
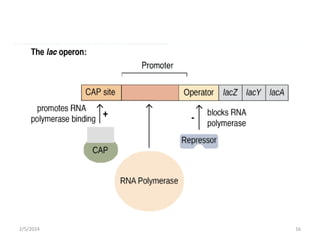
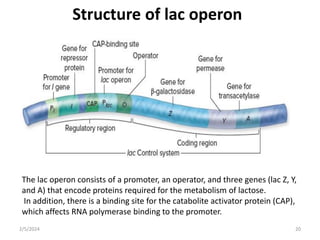
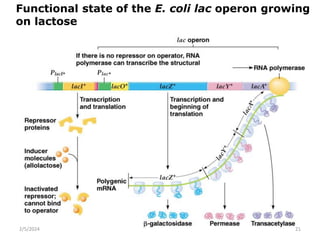
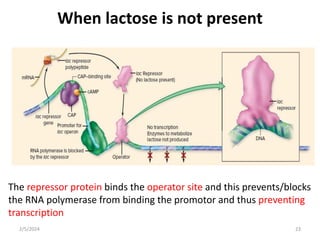
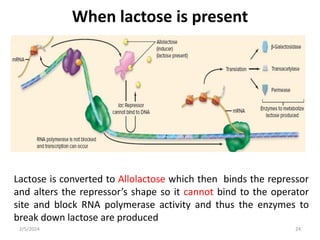
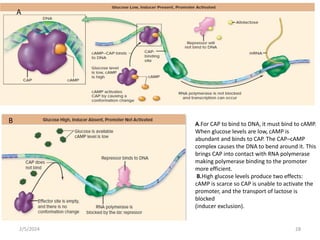
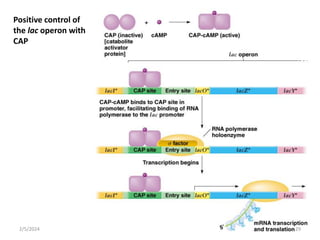
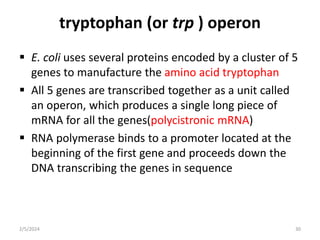
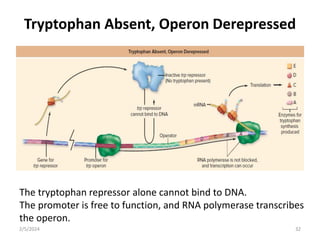
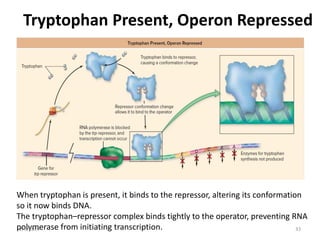
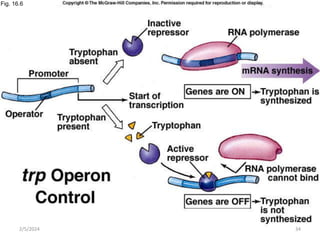
The document discusses gene regulation in prokaryotes. It explains that prokaryotes precisely regulate gene expression in response to environmental conditions by using operons. Operons allow groups of genes required for specific functions to be coordinately controlled. Examples like the lac and tryptophan operons in E. coli demonstrate how repressor proteins and small molecule effectors regulate operon expression depending on the availability of substrates like lactose and tryptophan. Quorum sensing is also discussed as a mechanism some bacteria use to regulate genes based on population density.