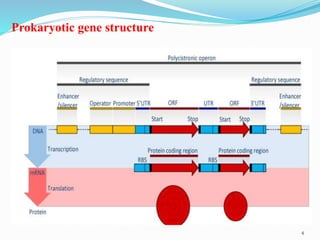
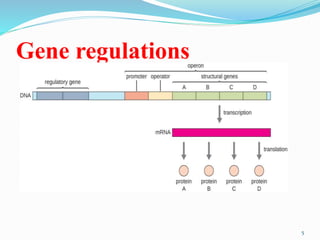
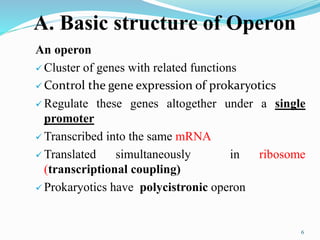
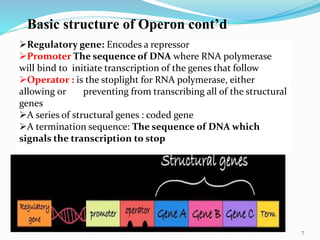
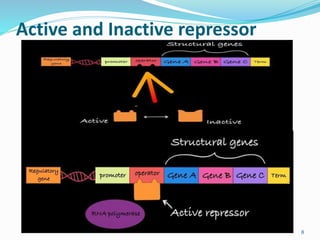
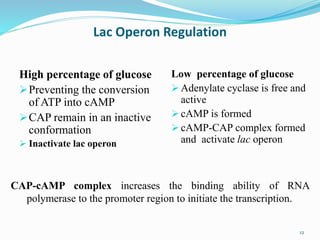
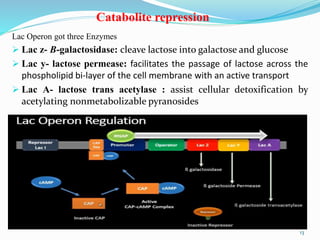
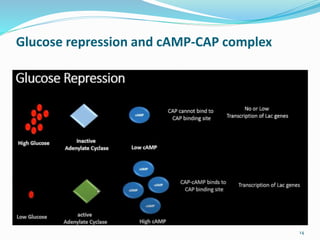
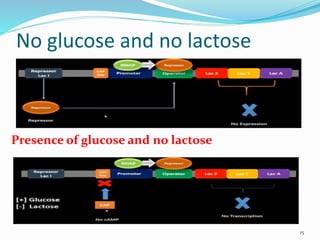
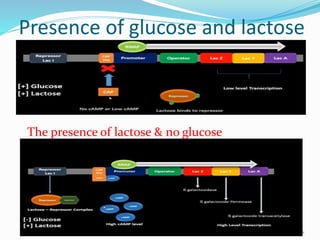
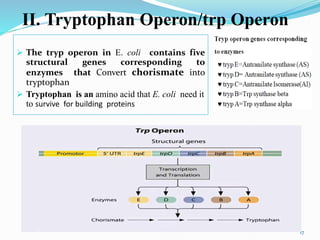
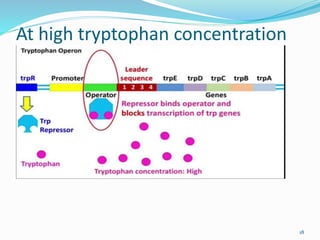
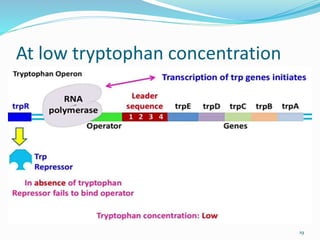
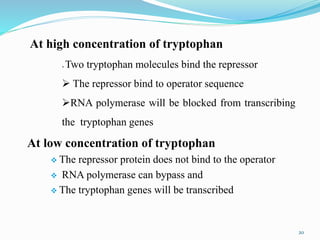
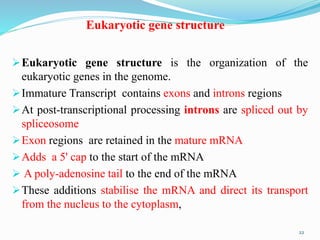
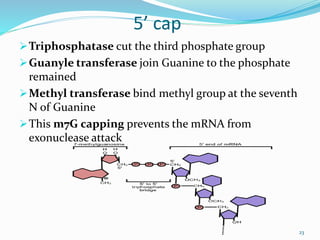
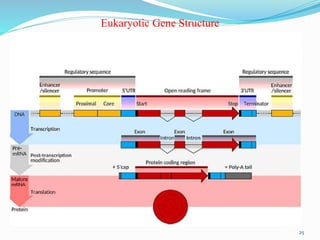
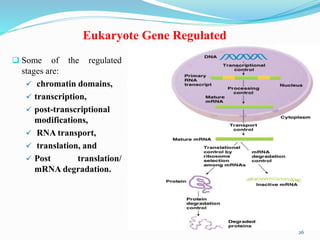
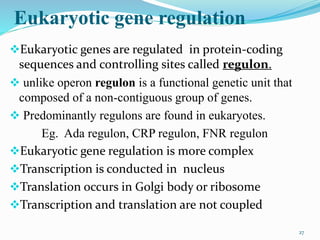
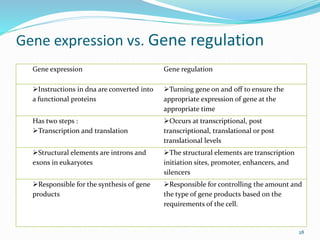
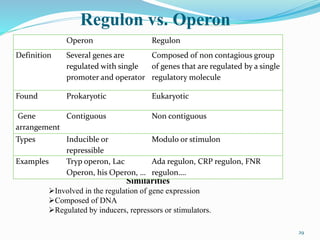
The document discusses prokaryotic and eukaryotic gene expression and regulation, including fundamental concepts such as operon structures and the mechanisms of gene regulation. It elaborates on specific operons like the lactose and tryptophan operons in prokaryotes, detailing their functions and regulatory factors. Additionally, it highlights the complexity of eukaryotic gene regulation, emphasizing processes such as RNA splicing and transport, along with differences between regulons and operons.