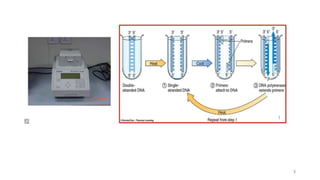
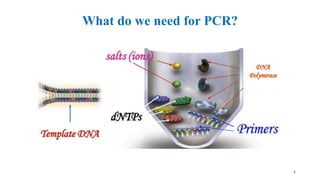
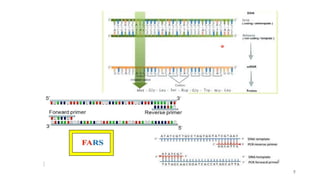
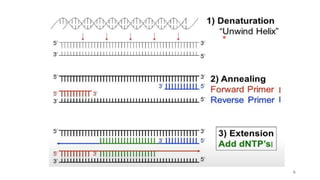
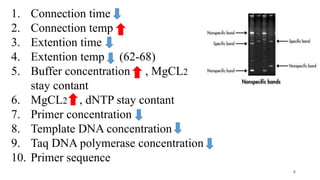
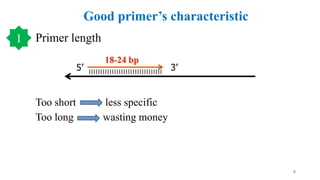
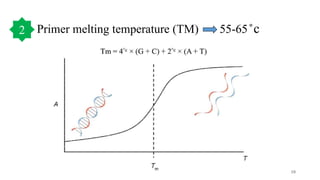
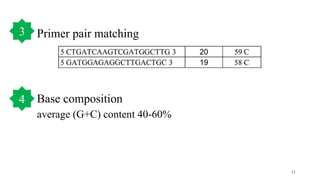
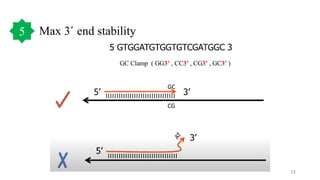
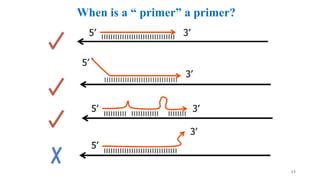
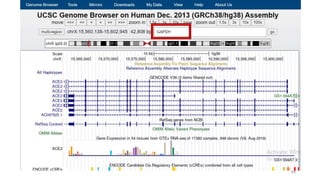
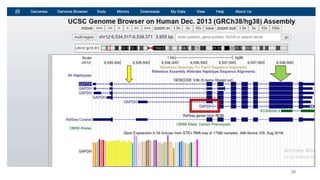
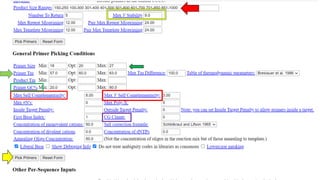
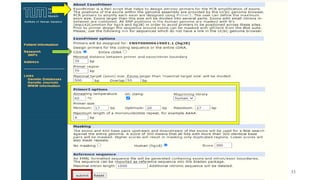
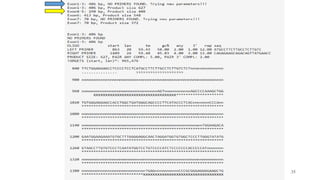
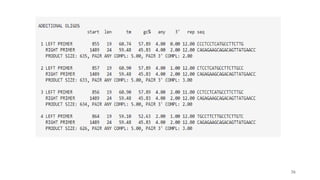
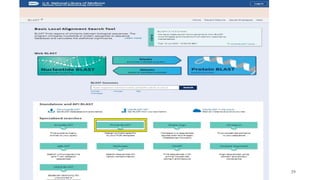
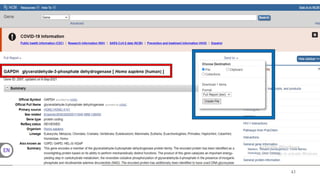
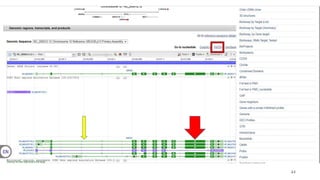
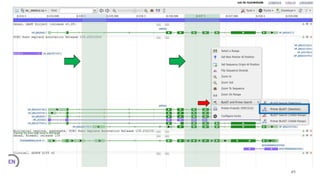
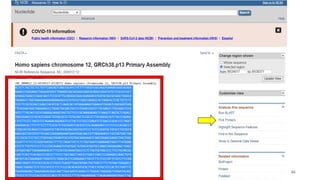
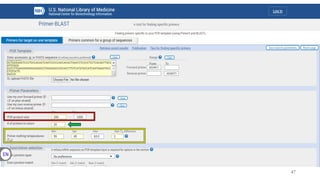
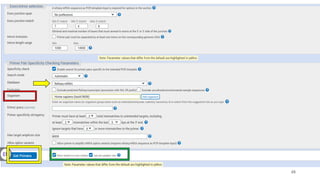
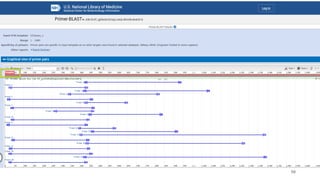
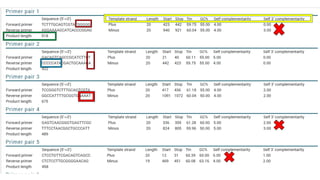
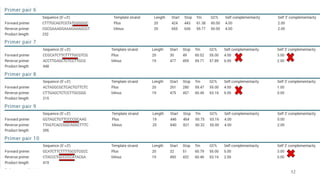
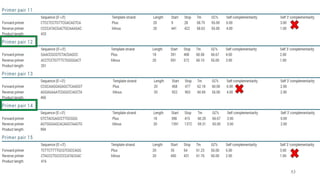
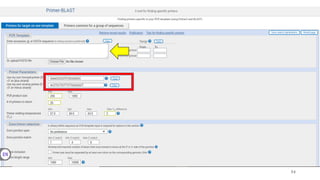
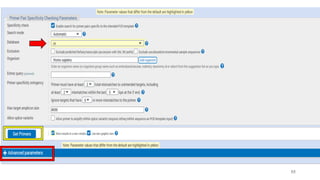
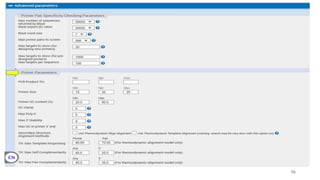
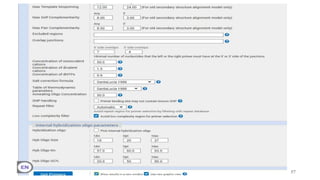
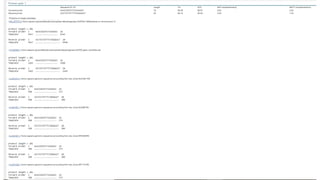
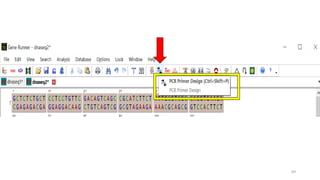
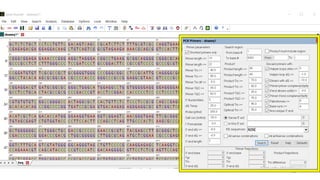
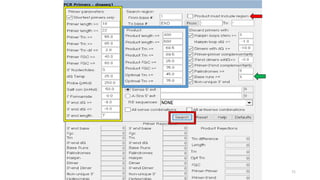
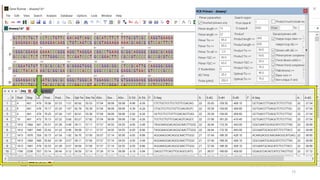
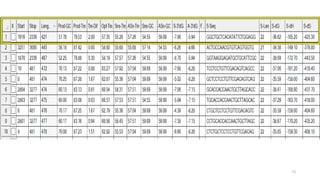
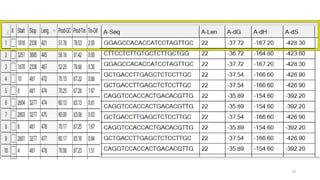
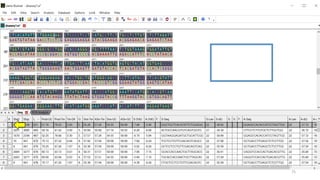
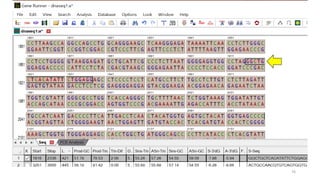
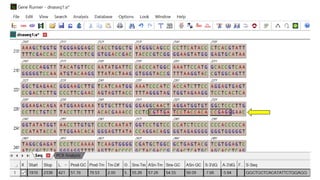
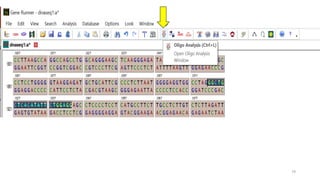
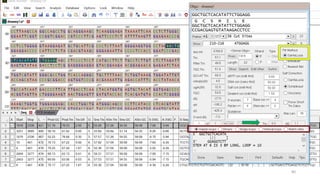
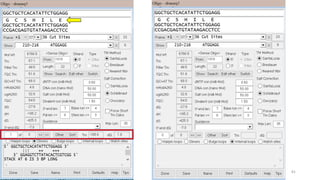
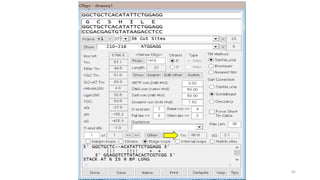
The document discusses PCR (polymerase chain reaction) and primer design. It explains that each PCR cycle contains 3 steps: denaturation, primer annealing, and primer extension. These cycles are usually repeated 25-40 times. It also discusses various factors that must be programmed and optimized for PCR, including connection time and temperature, extension time and temperature, buffer concentration, primer characteristics like length, melting temperature, sequence, and potential for hairpin formations or primer-dimer formations. The document emphasizes that primers should be designed to be specific and avoid these unwanted structures.