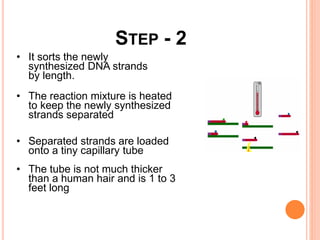
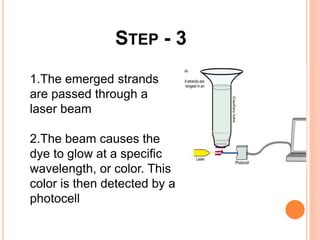
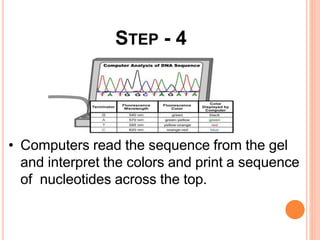
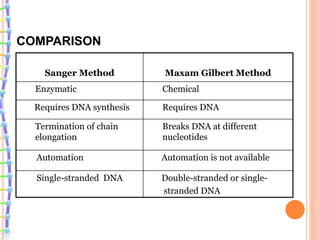
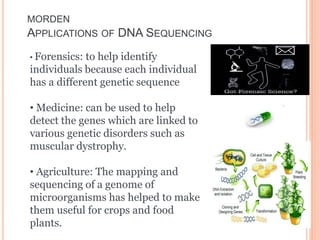
The document summarizes a presentation on bioinformatics and DNA sequencing. It includes 5 group members who each discuss an aspect of DNA sequencing. It describes how DNA stores genetic information and its structure. It then explains the history of DNA discovery and different sequencing methods, including the Sanger method. Modern applications of sequencing in forensics, medicine, and agriculture are outlined. The human genome project is summarized as a large international effort to sequence the entire human genome.