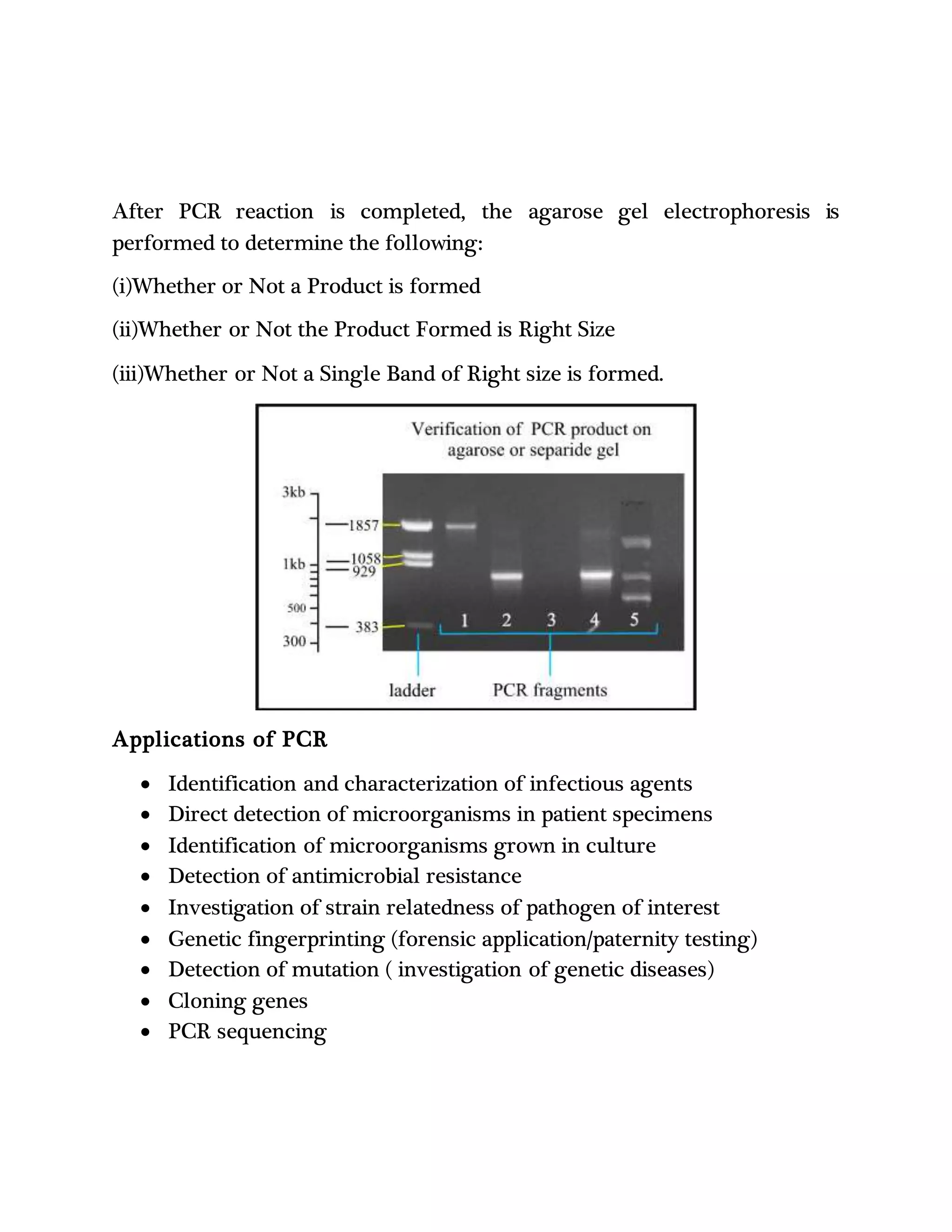
Polymerase chain reaction (PCR) is a technique used to amplify a single copy of a DNA segment into millions of copies. PCR uses repeated cycles of heating and cooling of the DNA sample to denature and separate the double-stranded DNA, followed by annealing of primers and extension of the DNA strands by a DNA polymerase. This allows the targeted DNA segment to be exponentially amplified. A basic PCR setup requires DNA template, primers, dNTPs, buffer solution, bivalent cations such as magnesium, and a thermostable DNA polymerase like Taq polymerase. PCR has applications in cloning genes, detecting genetic mutations and microorganisms, and genetic fingerprinting.