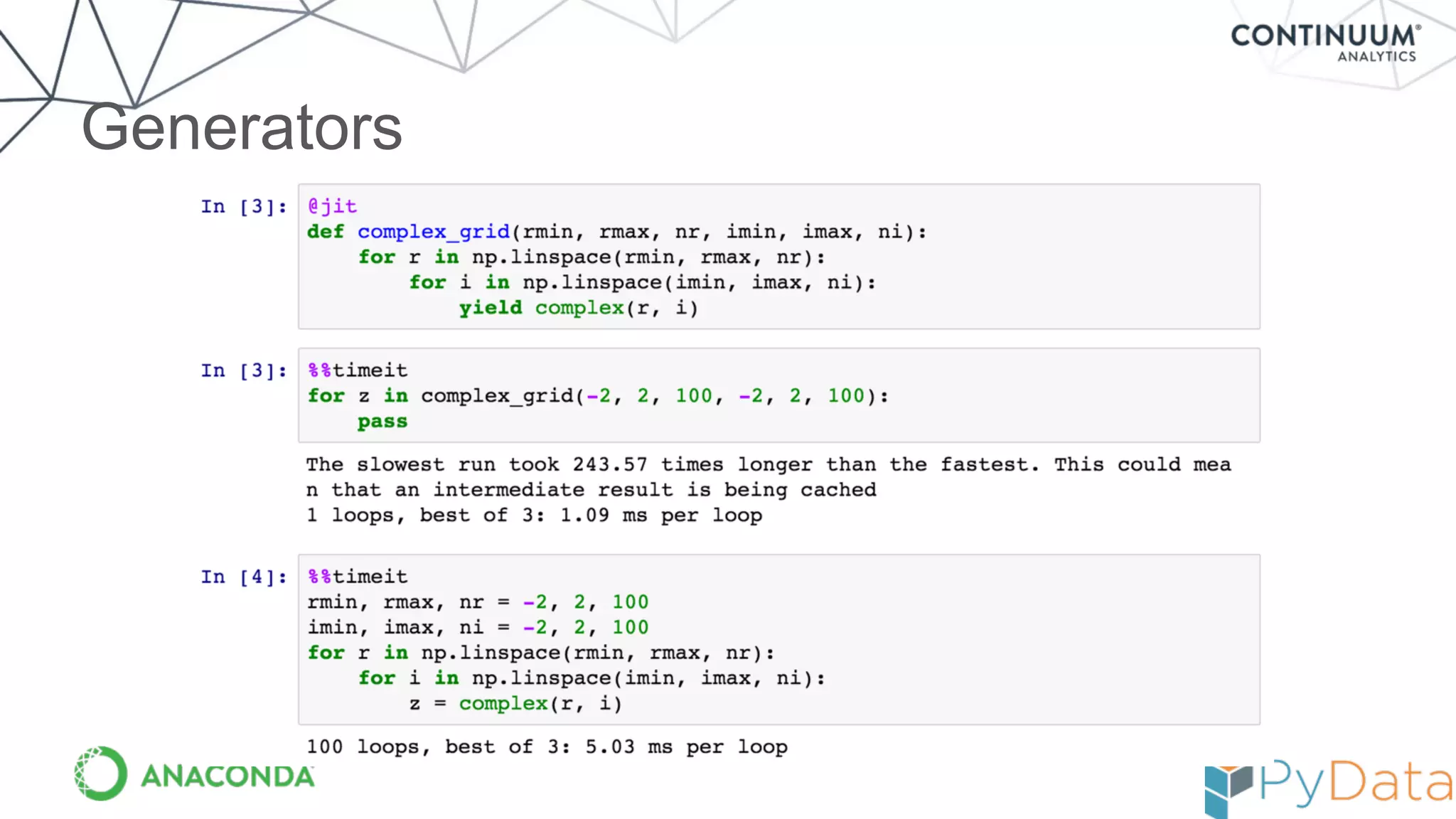
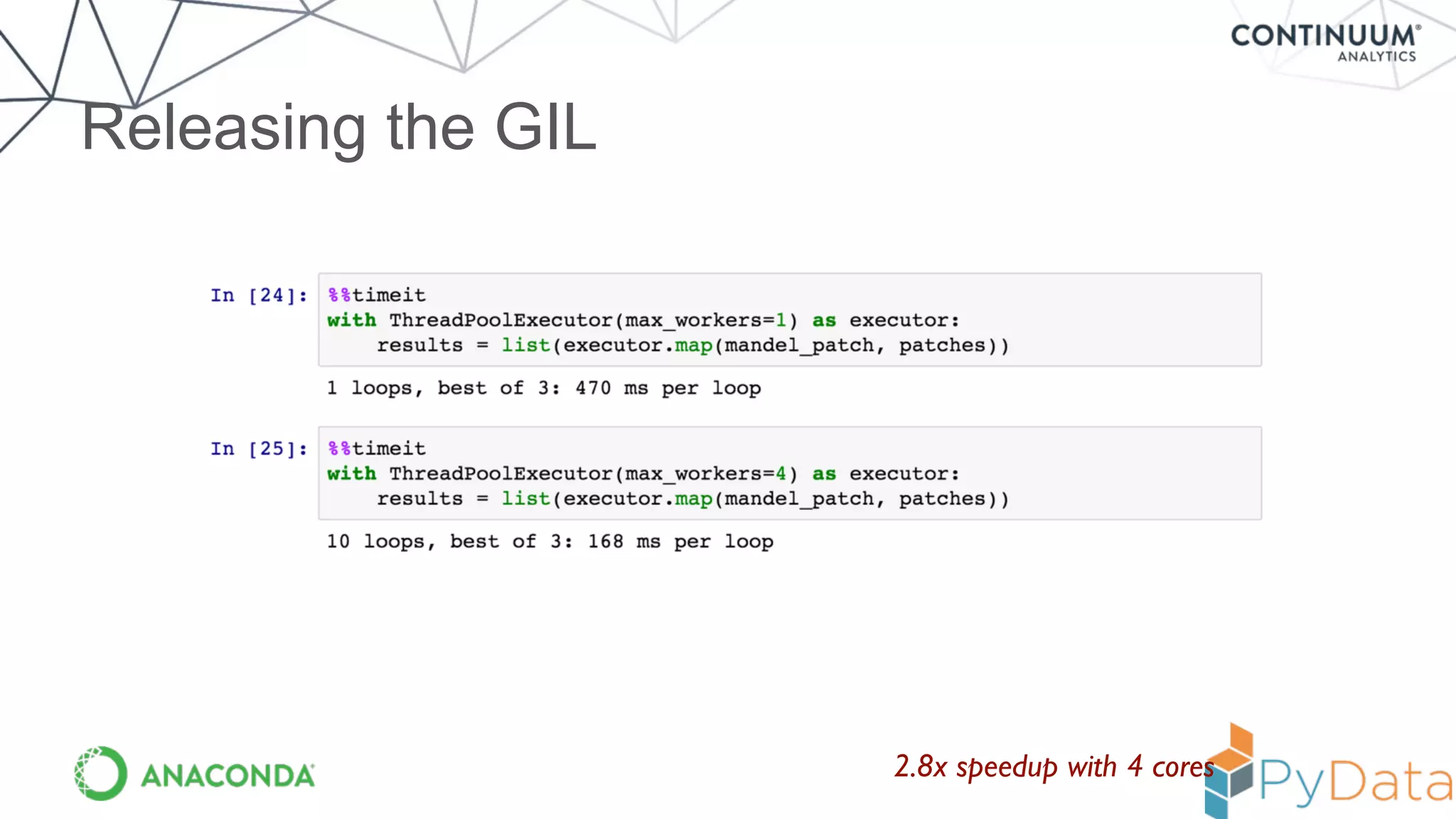
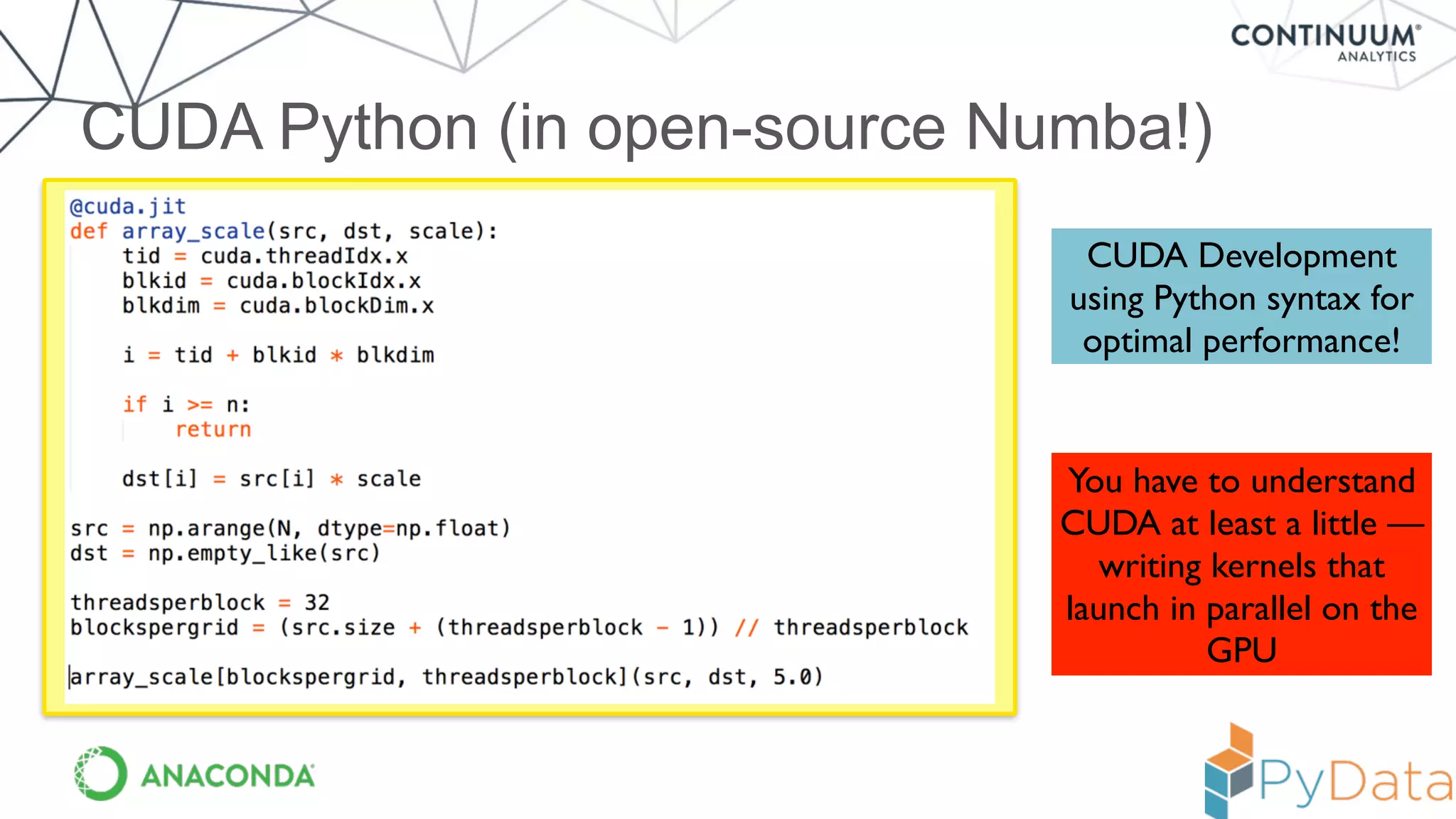
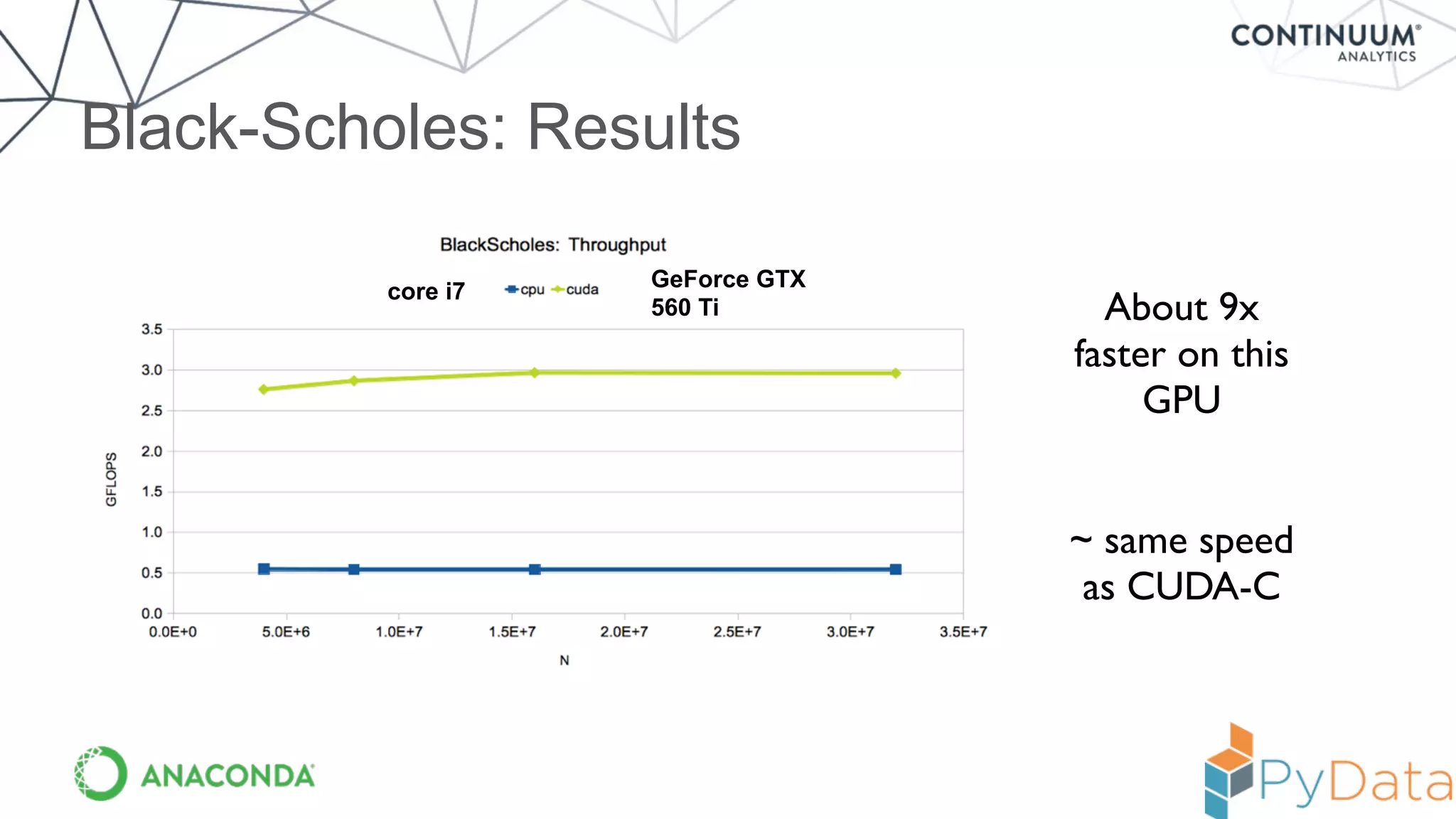
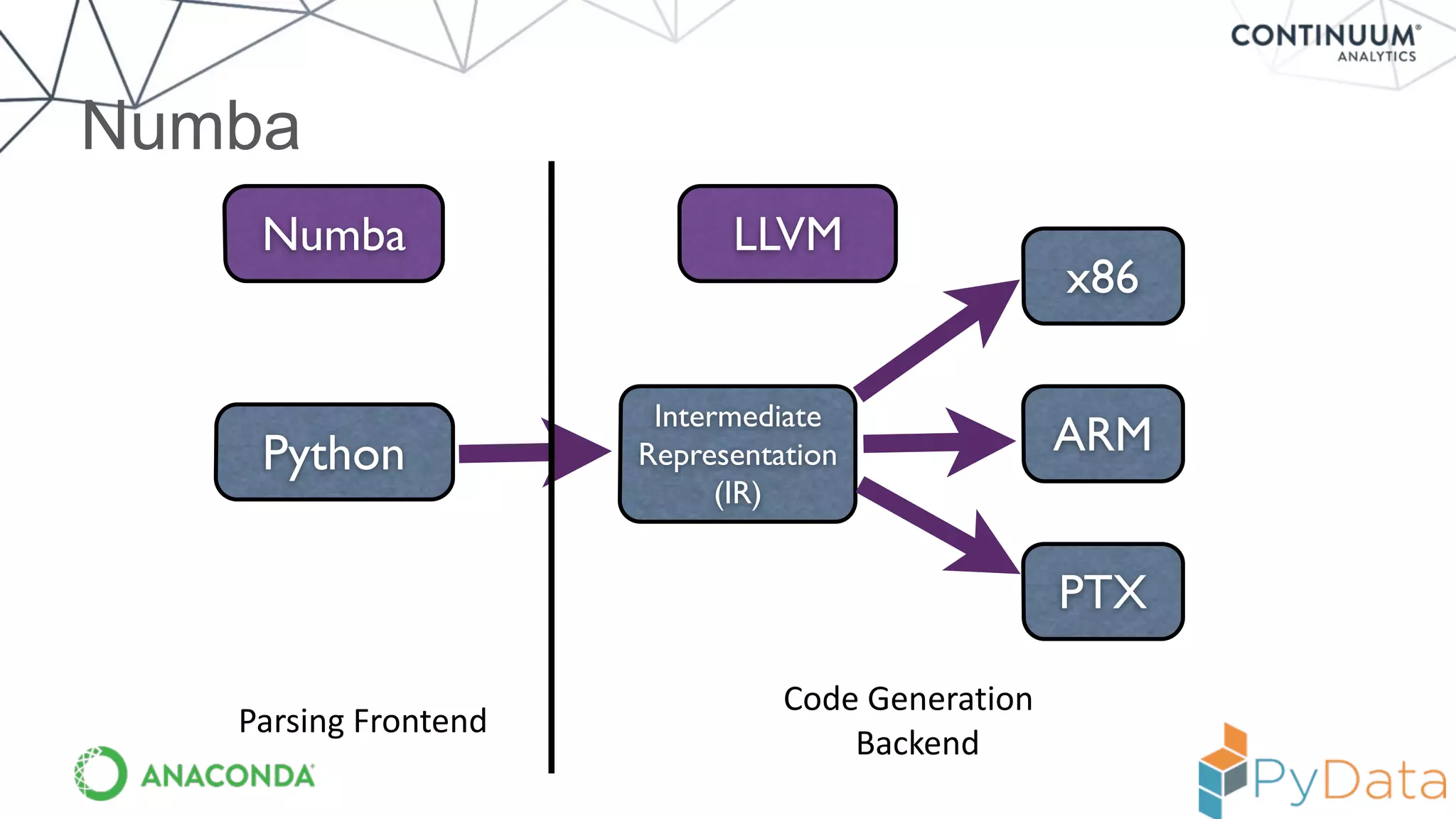
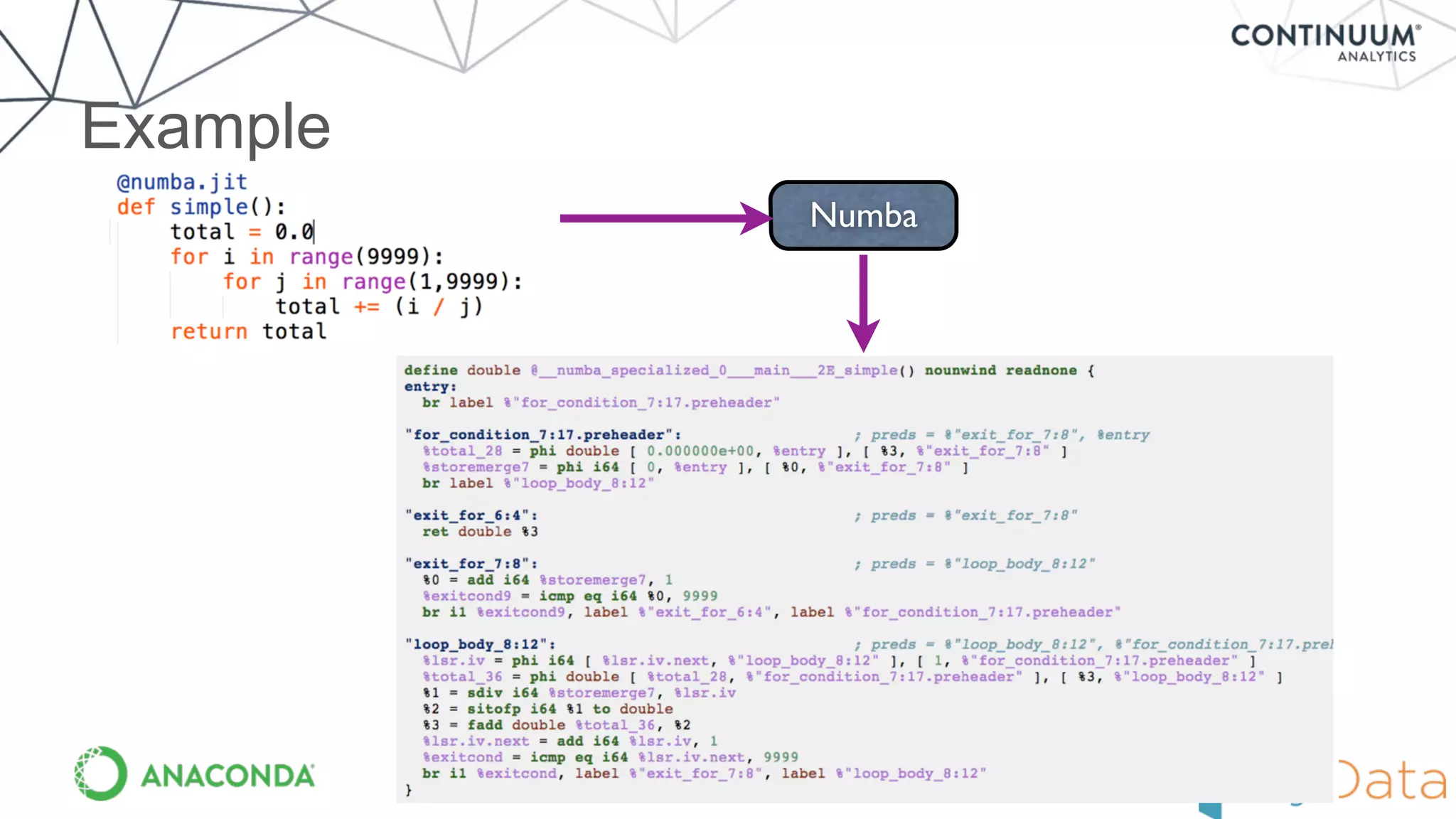
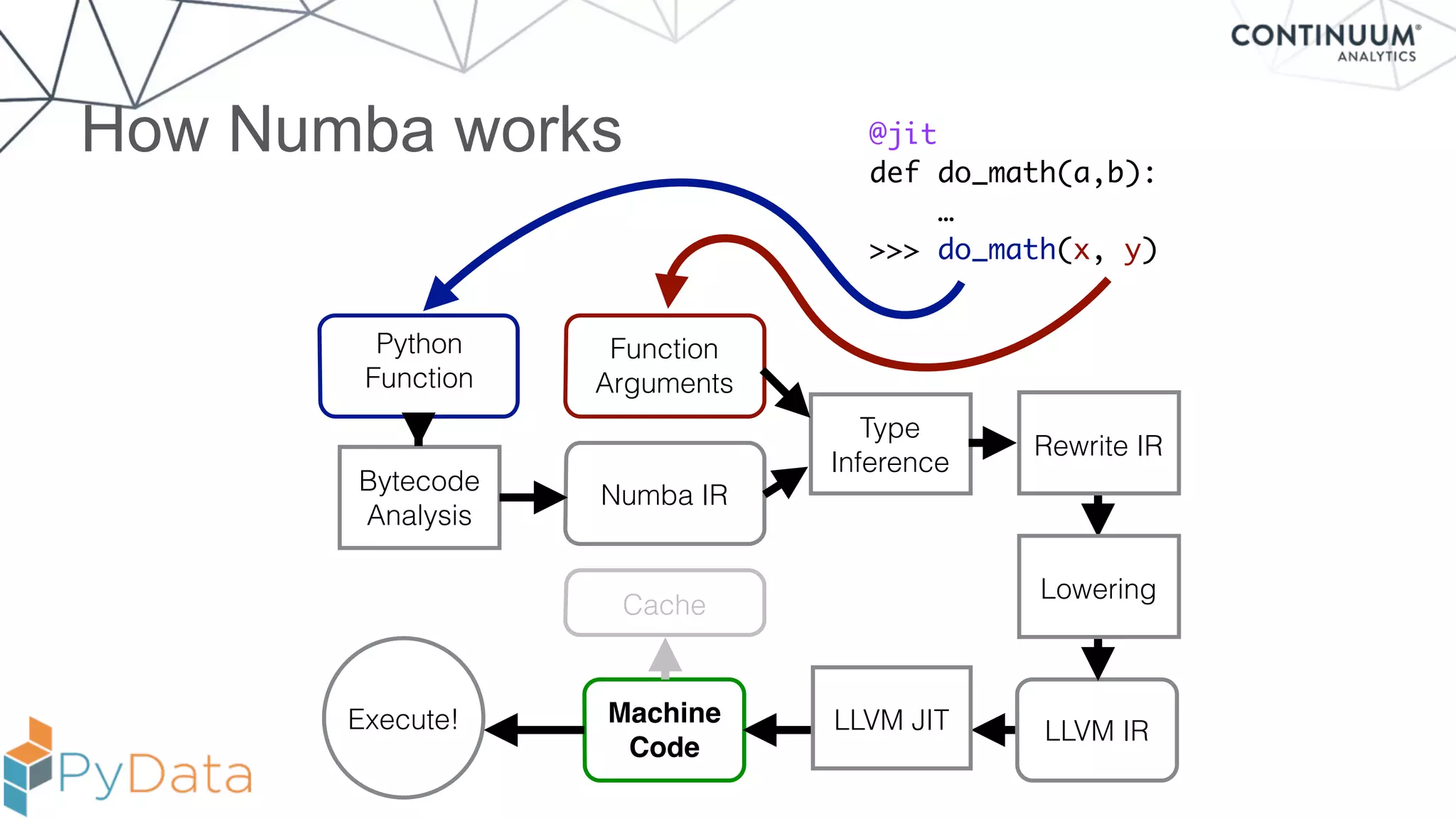
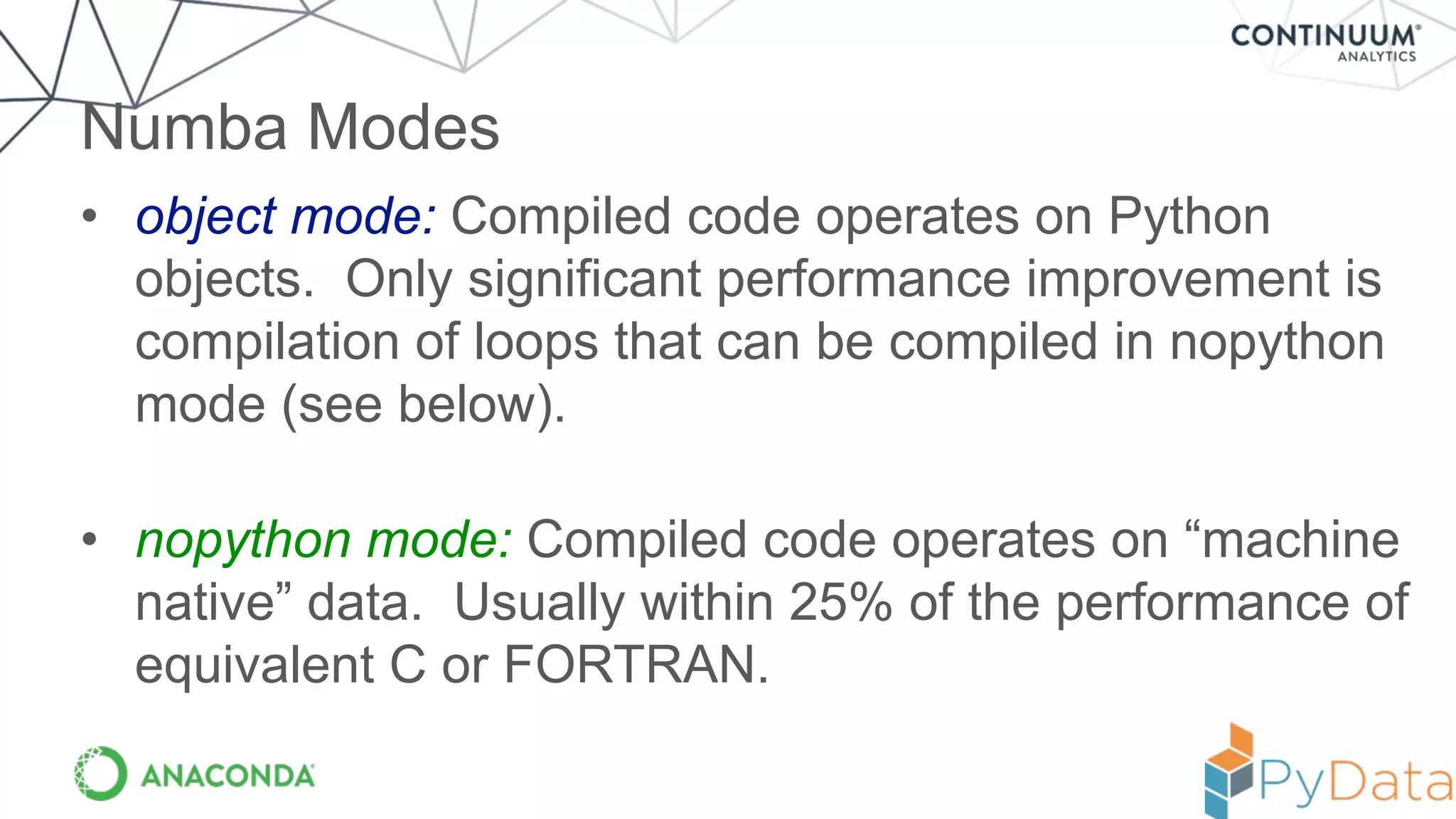
Numba provides a way to write high performance Python code using NumPy-like syntax. It works by compiling Python code with NumPy arrays and loops into fast machine code using the LLVM compiler. This allows code written in Python to achieve performance comparable to C/C++ with little or no code changes required. Numba supports CPU and GPU execution via backends like CUDA. It can improve performance of numerical Python code with features like releasing the global interpreter lock during compilation.





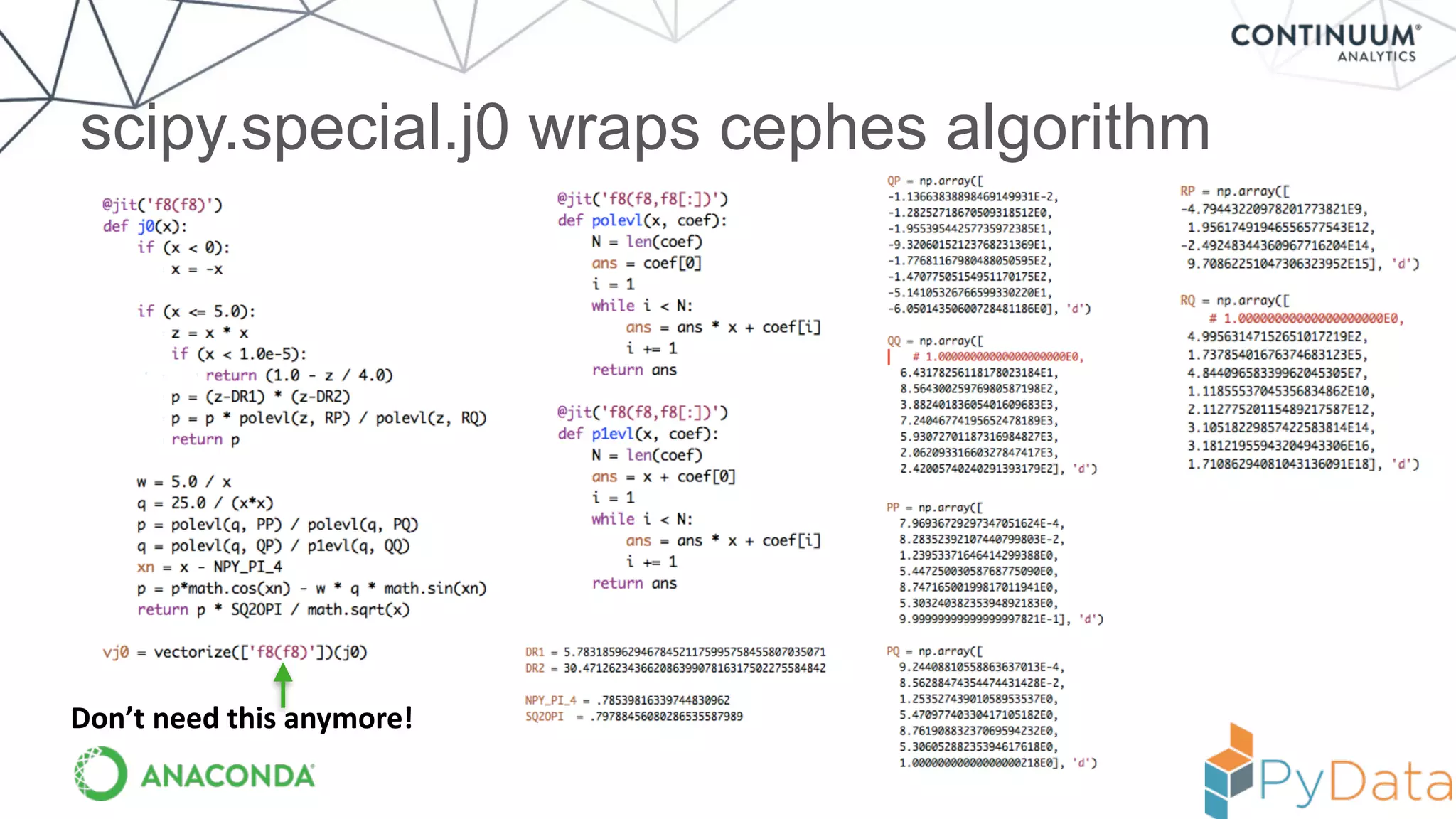
![Result --- equivalent to compiled code
In [6]: %timeit vj0(x)
10000 loops, best of 3: 75 us per loop
In [7]: from scipy.special import j0
In [8]: %timeit j0(x)
10000 loops, best of 3: 75.3 us per loop
But! Now code is in Python and can be experimented with
more easily (and moved to the GPU / accelerator more easily)!](https://image.slidesharecdn.com/numbaupdatejuly2015-150810154243-lva1-app6892/75/Numba-Flexible-analytics-written-in-Python-with-machine-code-speeds-and-avoiding-the-GIL-Travis-Oliphant-19-2048.jpg)