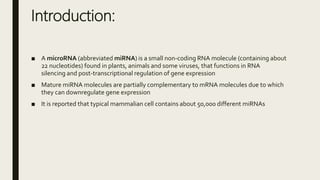
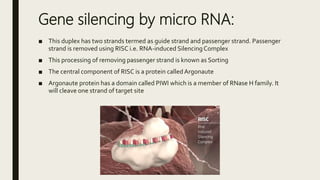
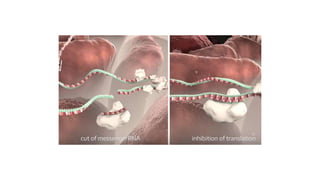
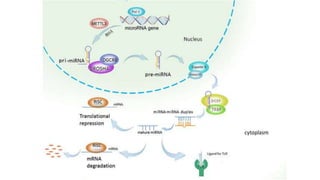
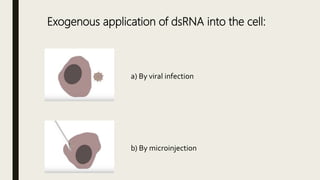
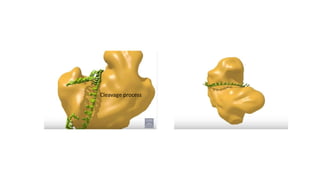
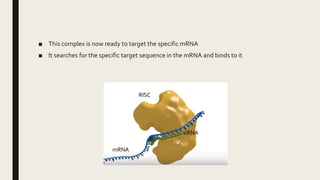
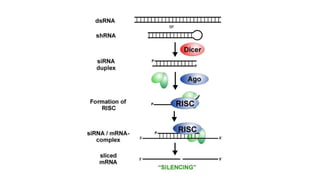
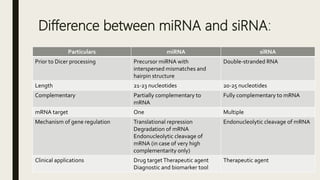
miRNA and siRNA and their application in crop improvement is a report submitted to Dr. Arna Das that discusses the history, biosynthesis, and mechanisms of gene silencing by miRNA and siRNA. It notes that miRNA is partially complementary to mRNA and downregulates gene expression, while siRNA is fully complementary and leads to mRNA degradation. The document outlines several applications of miRNA and siRNA in crop improvement, including increasing photosynthesis and yield in rice by modifying plant architecture, reducing lignin in plants, enhancing disease resistance, and prolonging fruit shelf life.