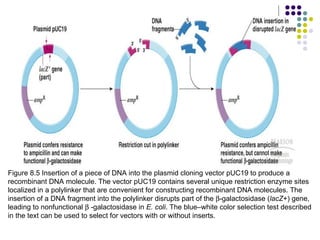
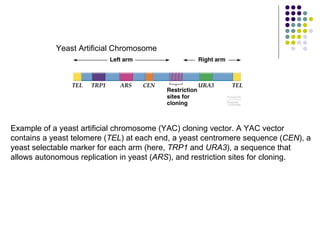
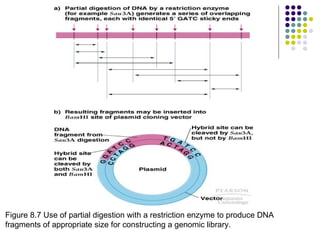
The document provides an overview of recombinant DNA technology and cloning techniques. It discusses:
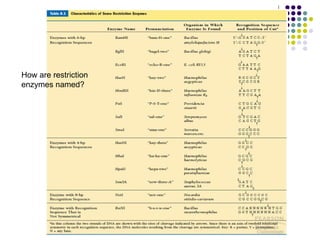
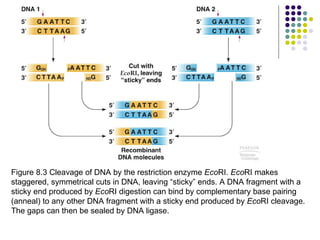
1) The general steps to clone DNA - isolating DNA from an organism, cutting it with restriction enzymes to create recombinant DNA, and introducing the DNA into a host.
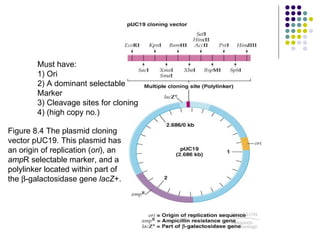
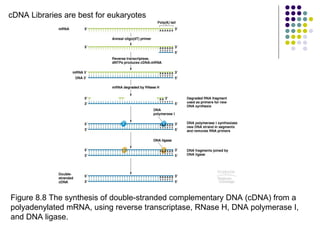
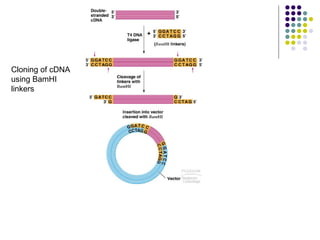
2) Types of cloning vectors like plasmids, artificial chromosomes, and viruses that are used to clone DNA fragments. Genomic and cDNA libraries containing clones of all DNA sequences are also described.
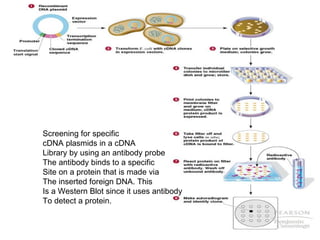
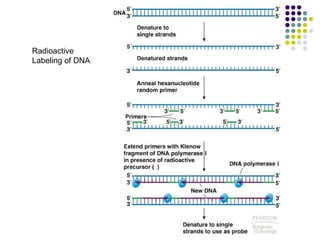
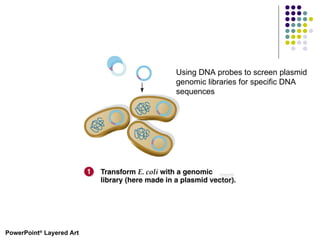
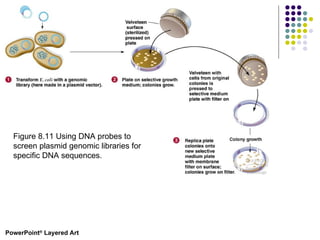
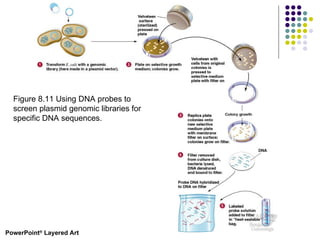
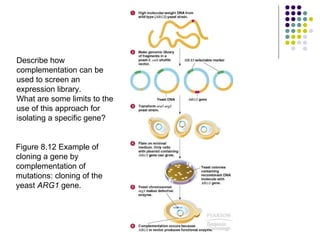
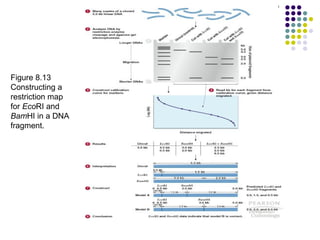
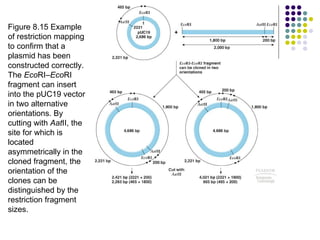
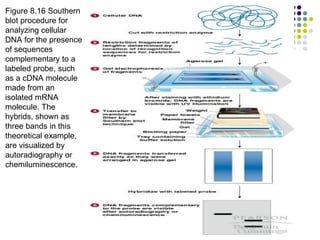
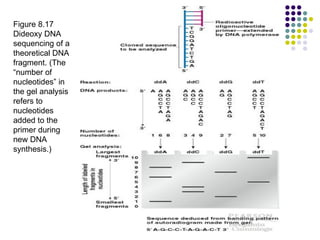
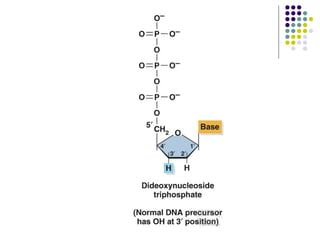
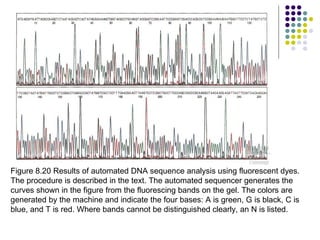
3) Techniques for identifying recombinant clones like hybridization probes, complementation of mutations, restriction mapping, and sequencing.