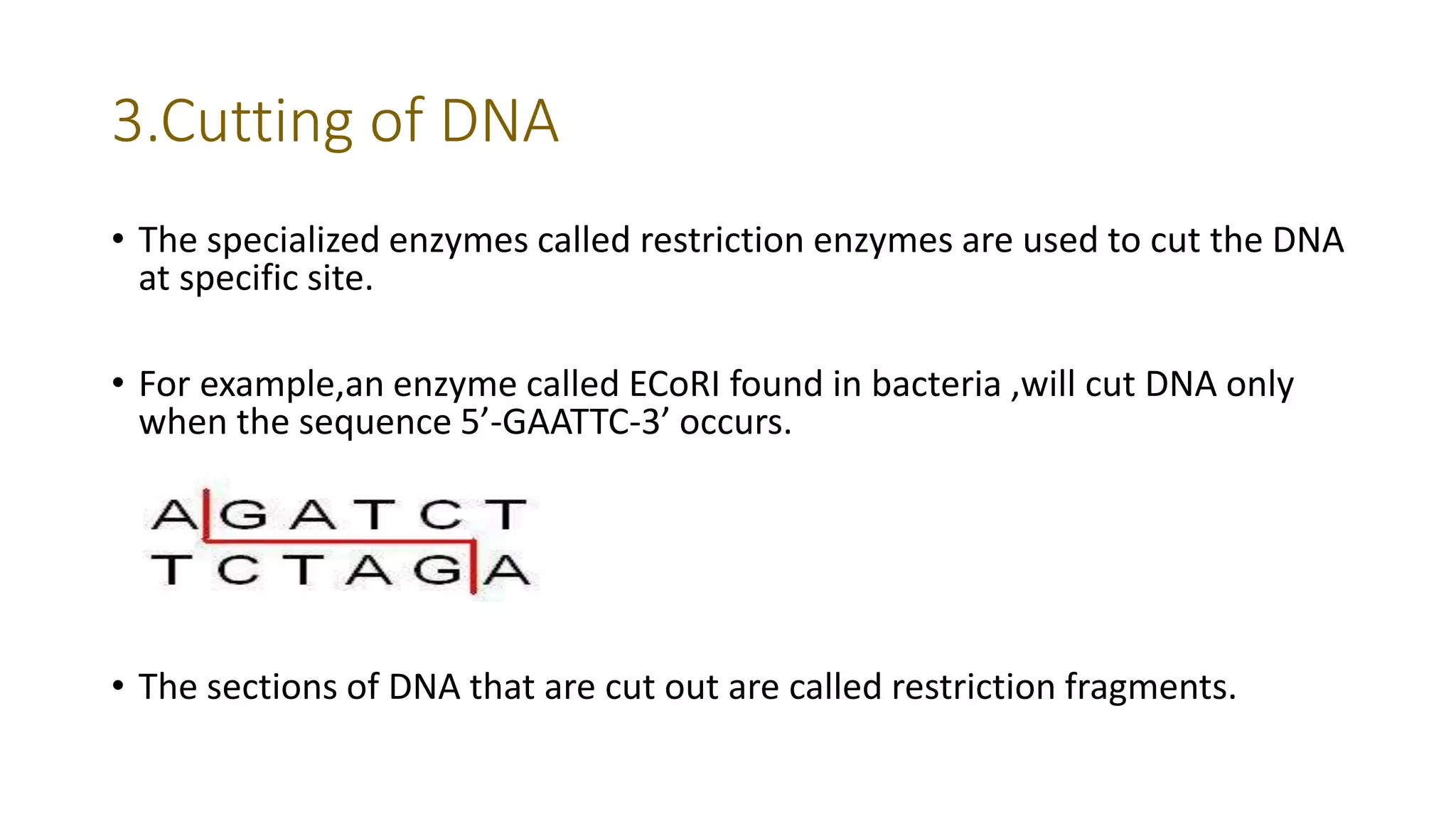
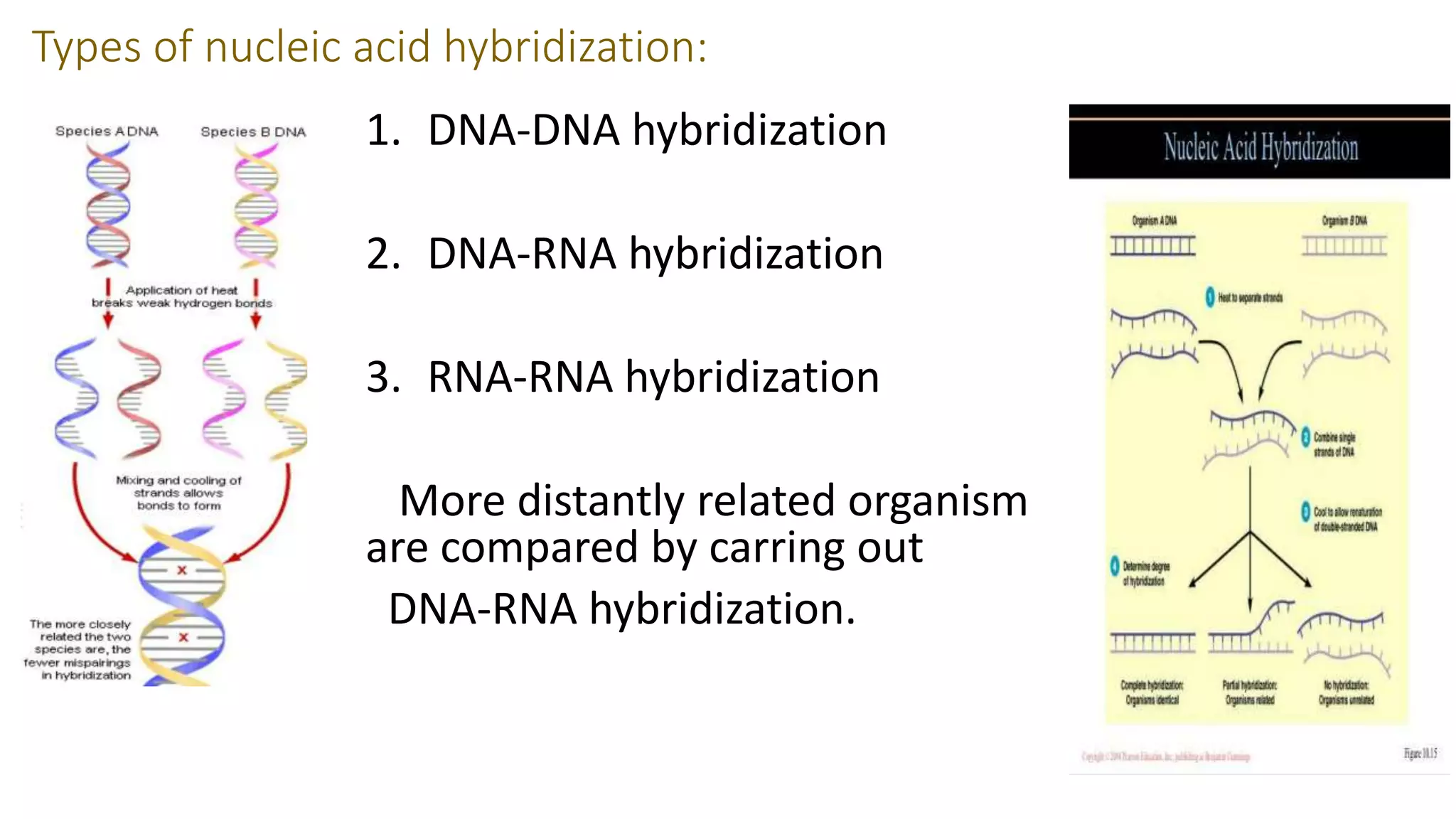
The document discusses DNA fingerprinting, nucleic acid hybridization, and rRNA sequence analysis. It defines DNA fingerprinting as a technique used to identify individuals by extracting and identifying their unique DNA base pair pattern. Nucleic acid hybridization is described as the process of forming a double stranded nucleic acid from joining two complementary single strands of DNA or RNA. rRNA sequence analysis is used to construct phylogenetic trees of life based on comparing 16s rRNA sequences between organisms.