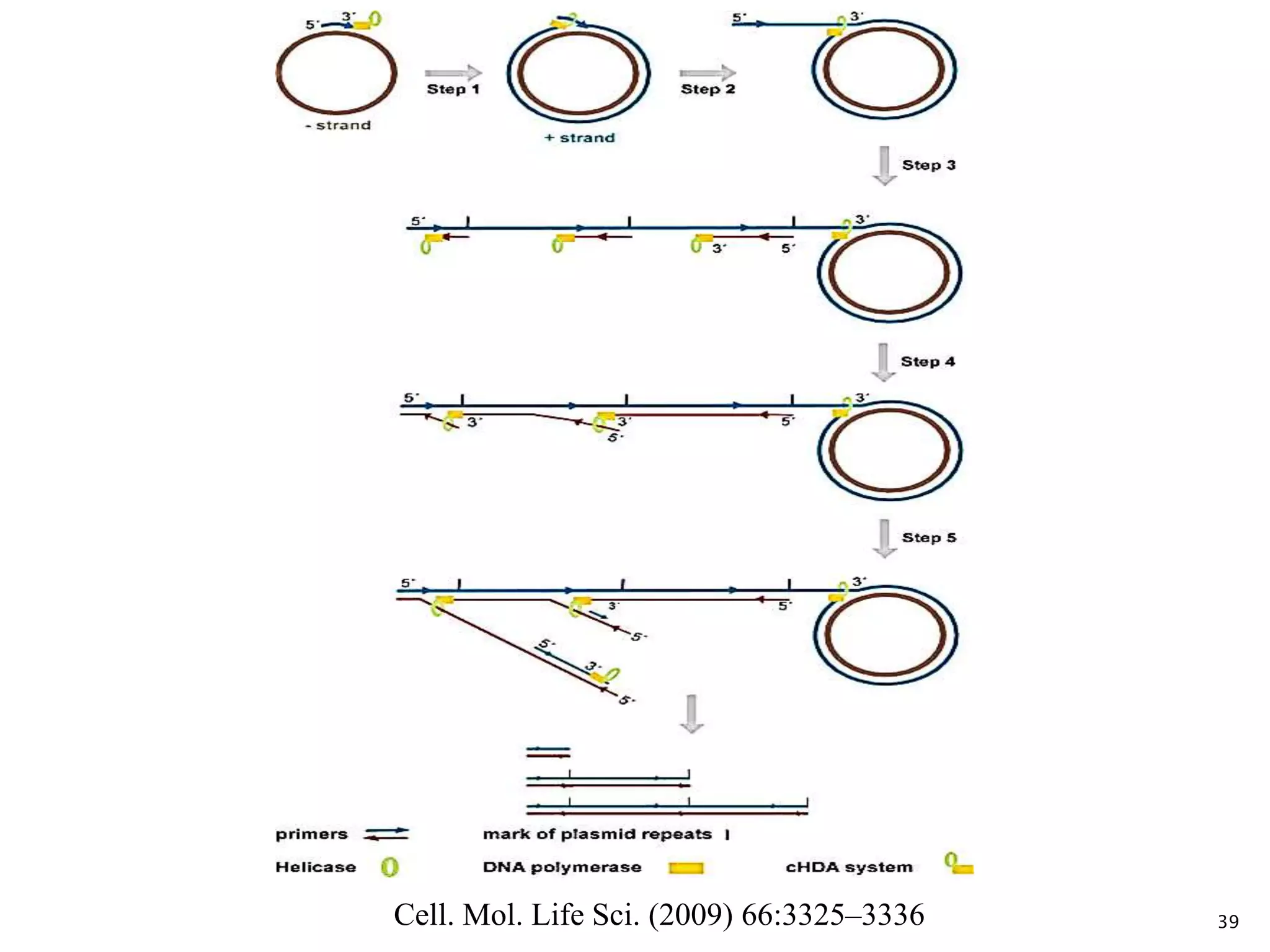
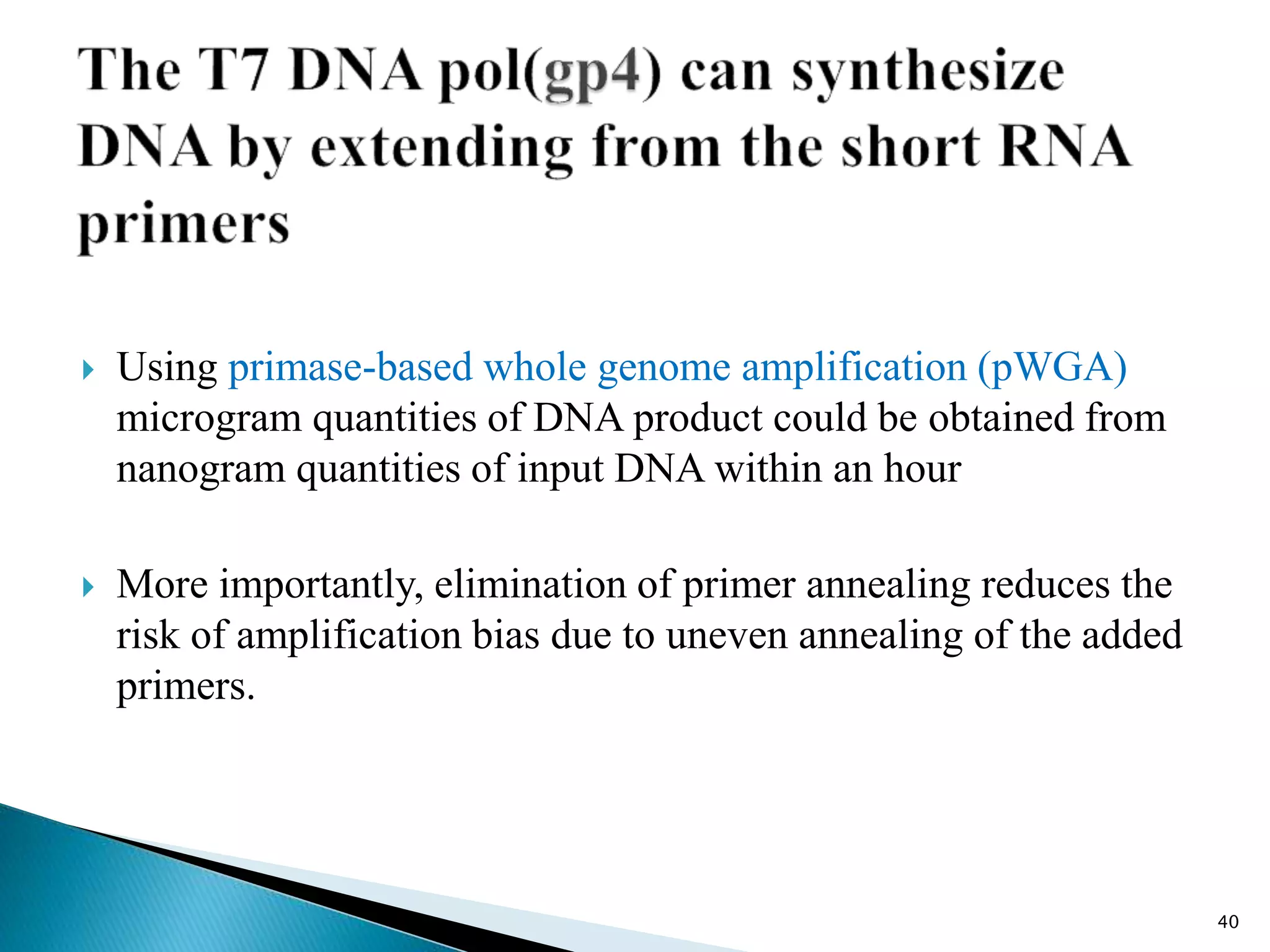
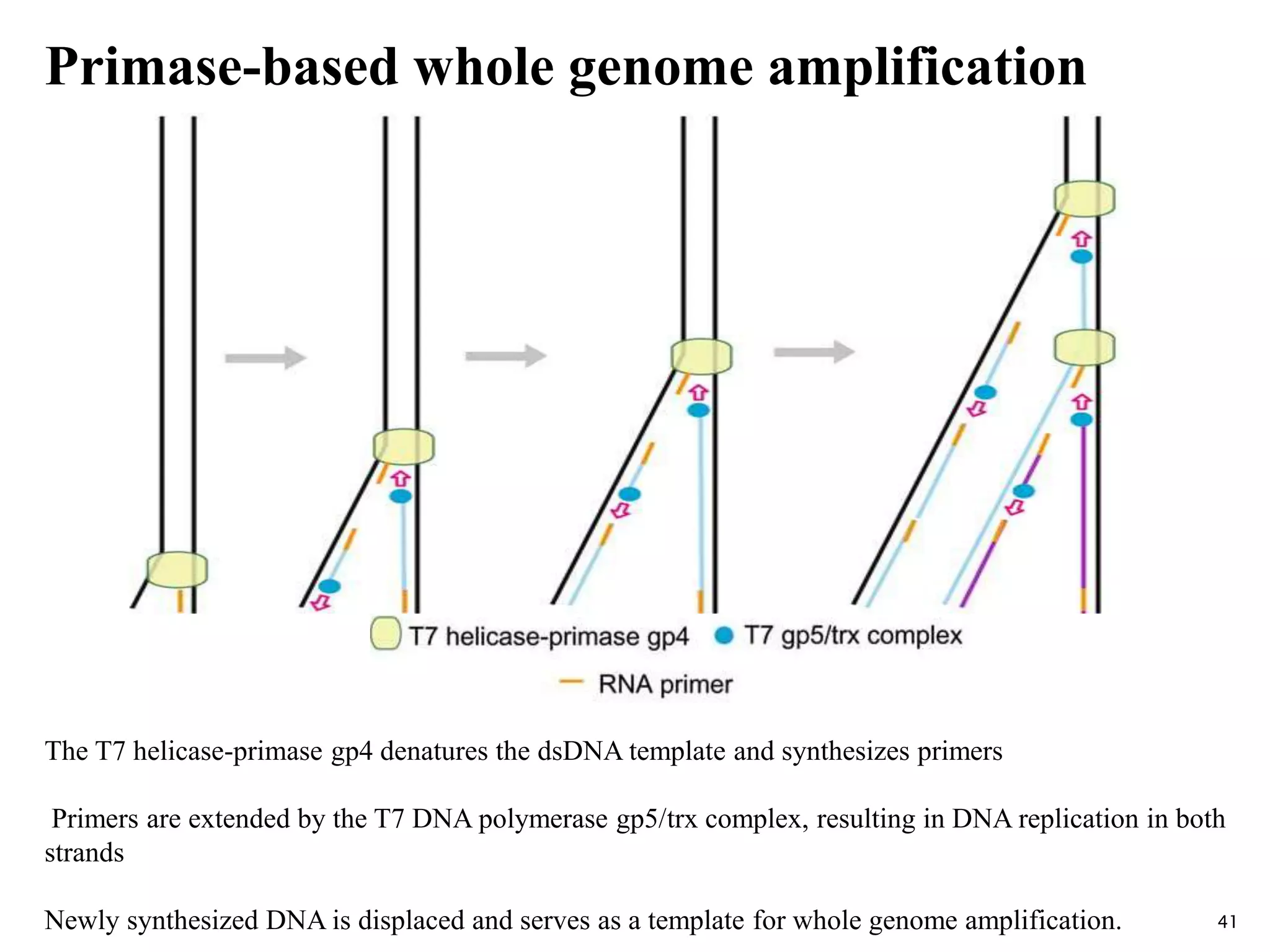
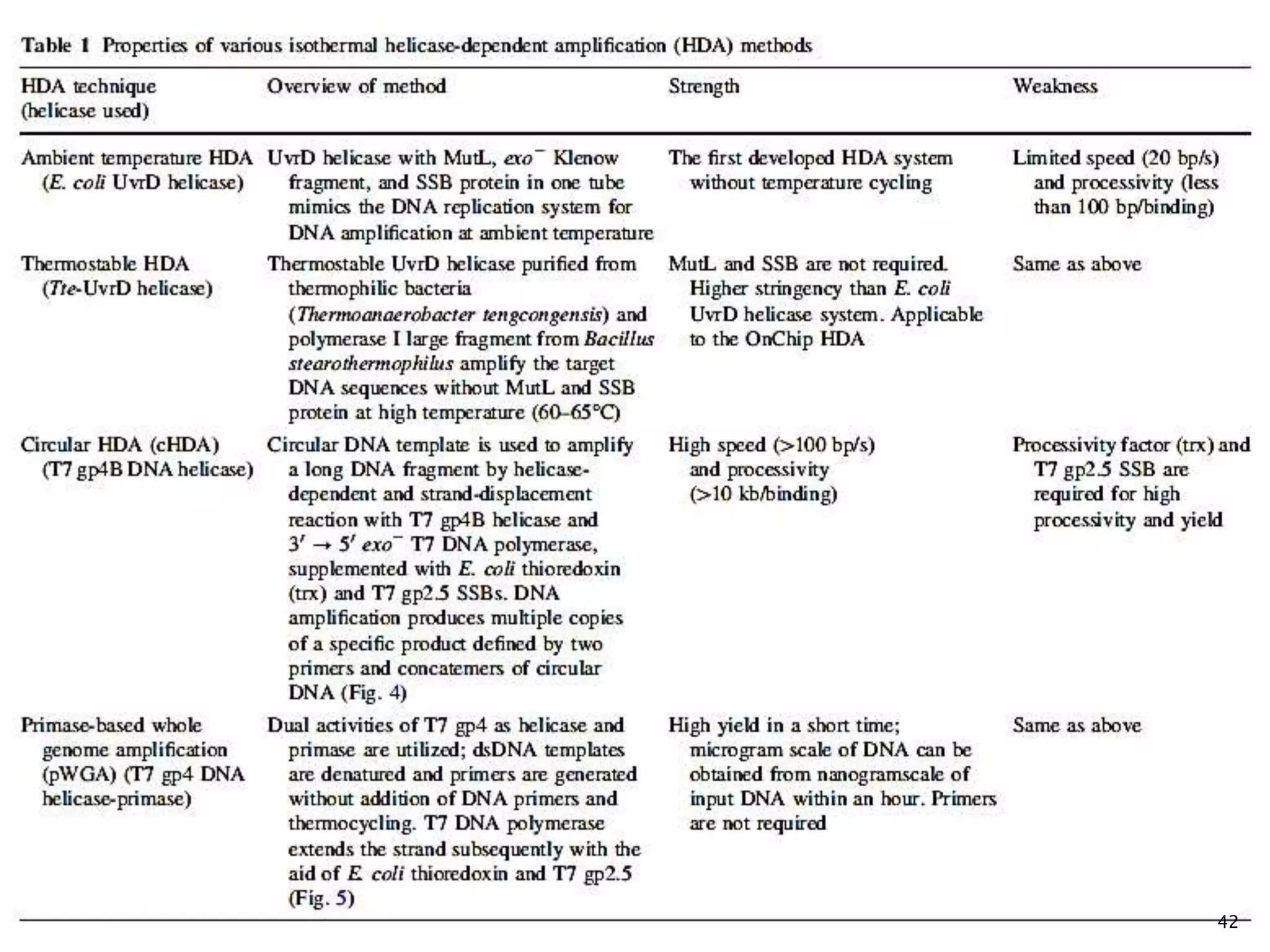
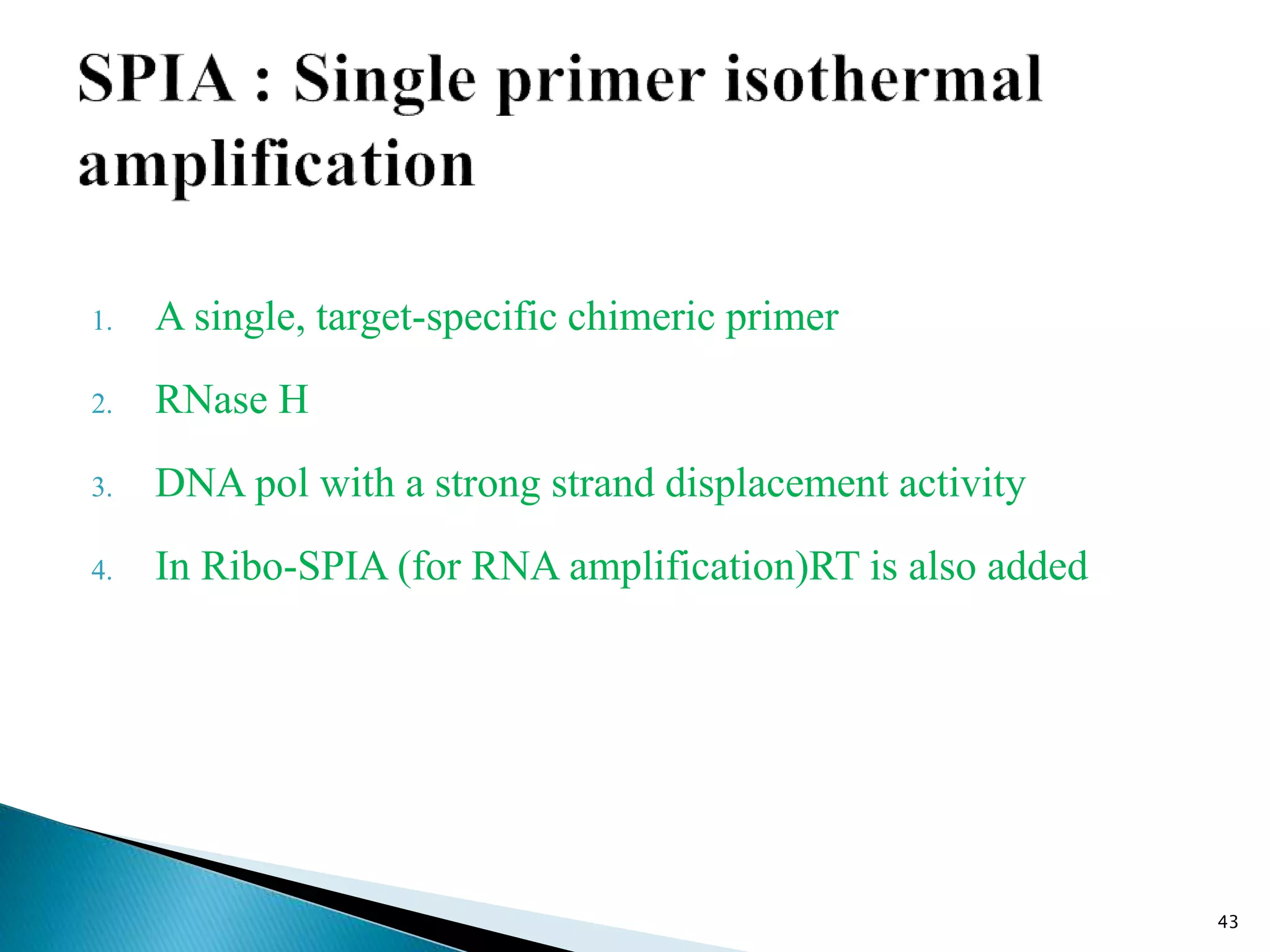
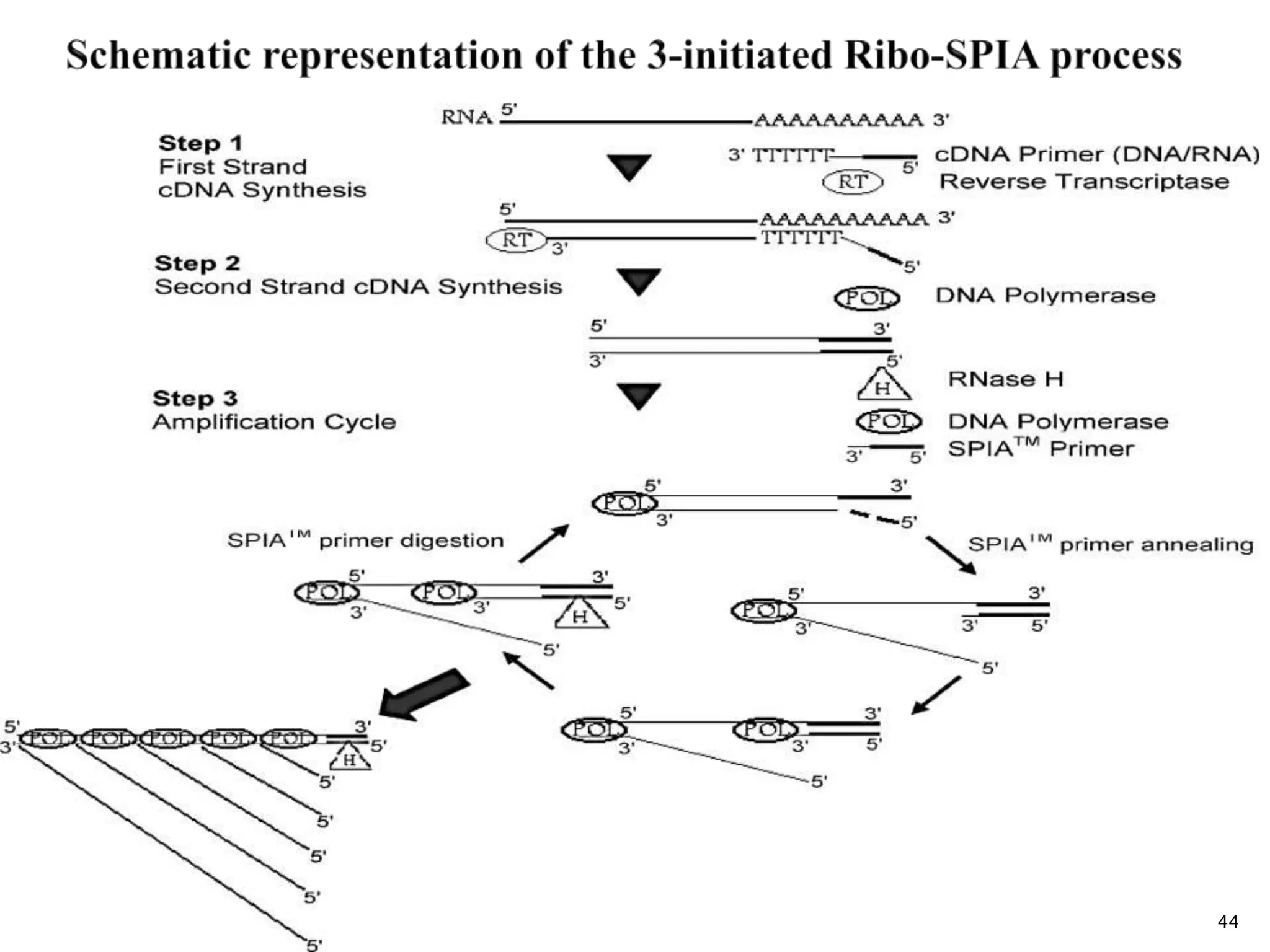
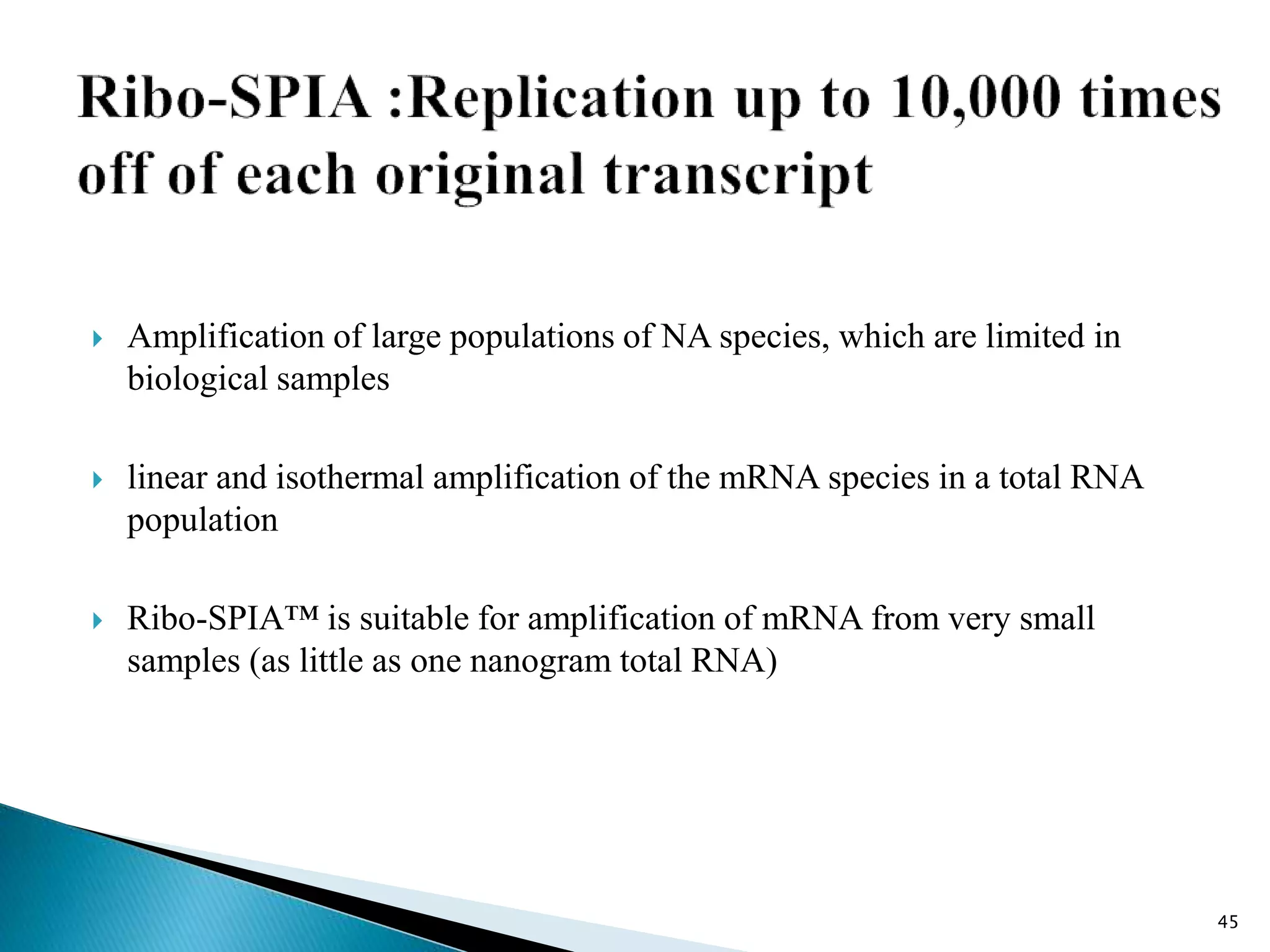
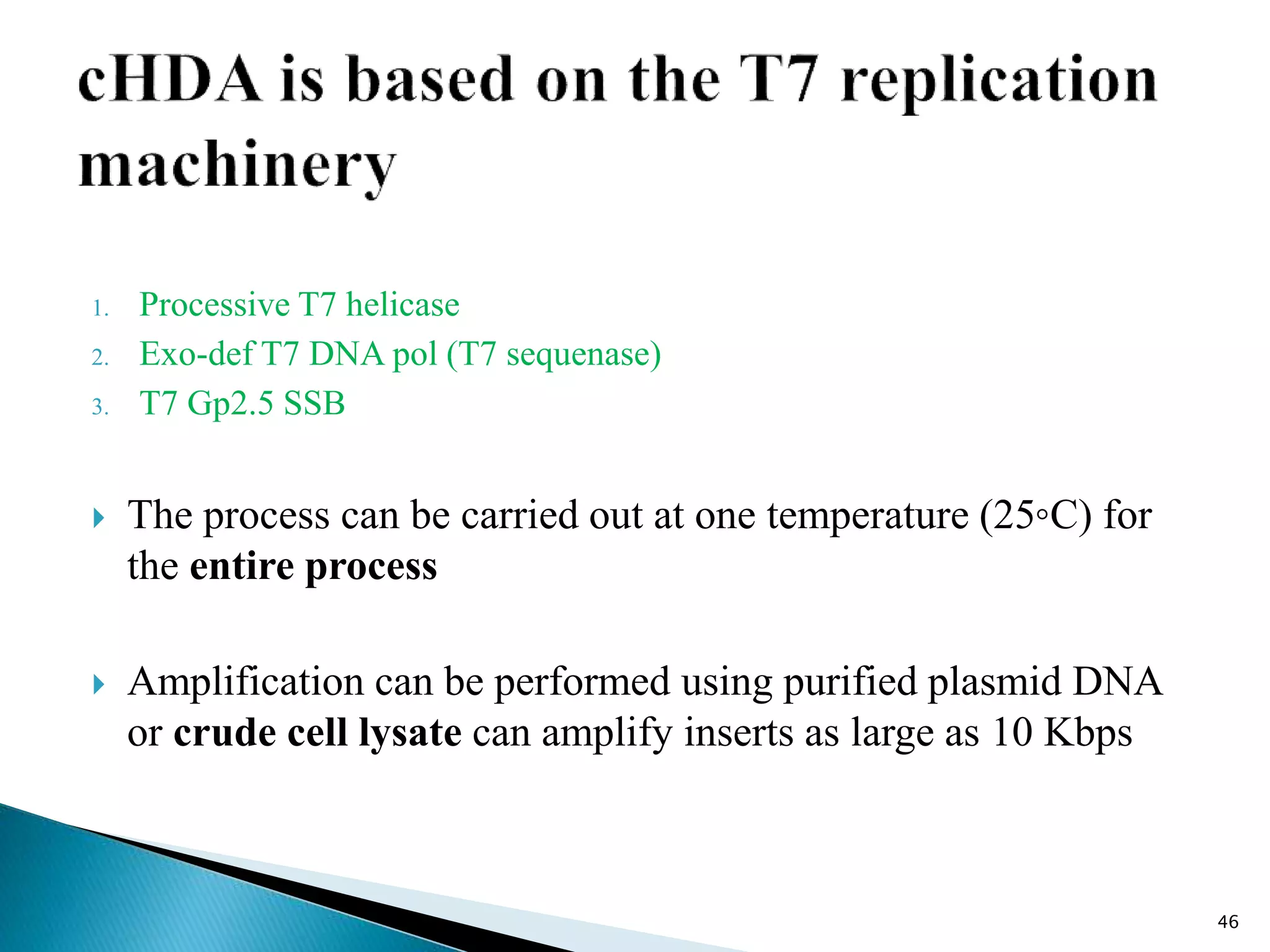
This document provides an overview of various isothermal nucleic acid amplification techniques, including:
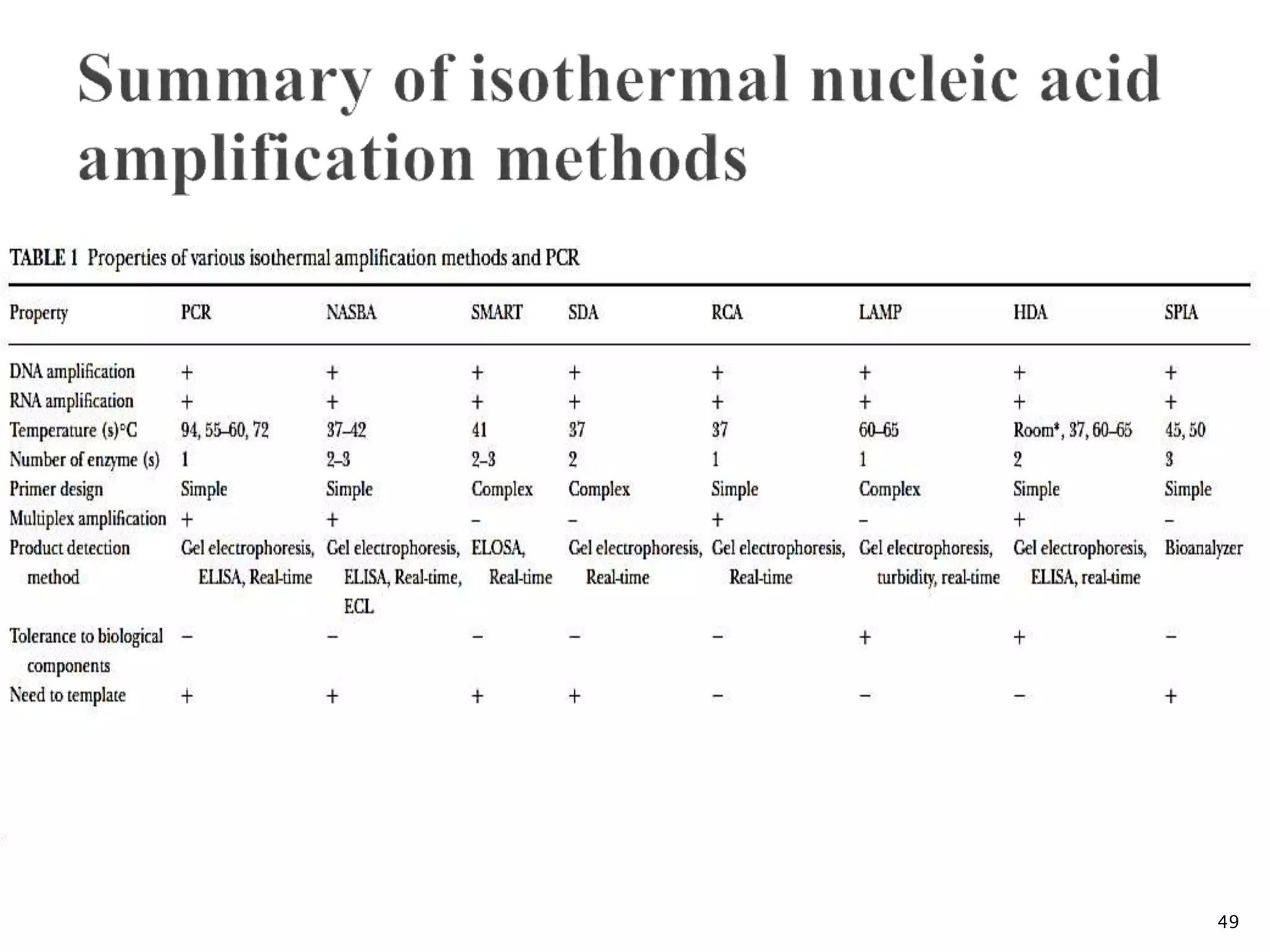
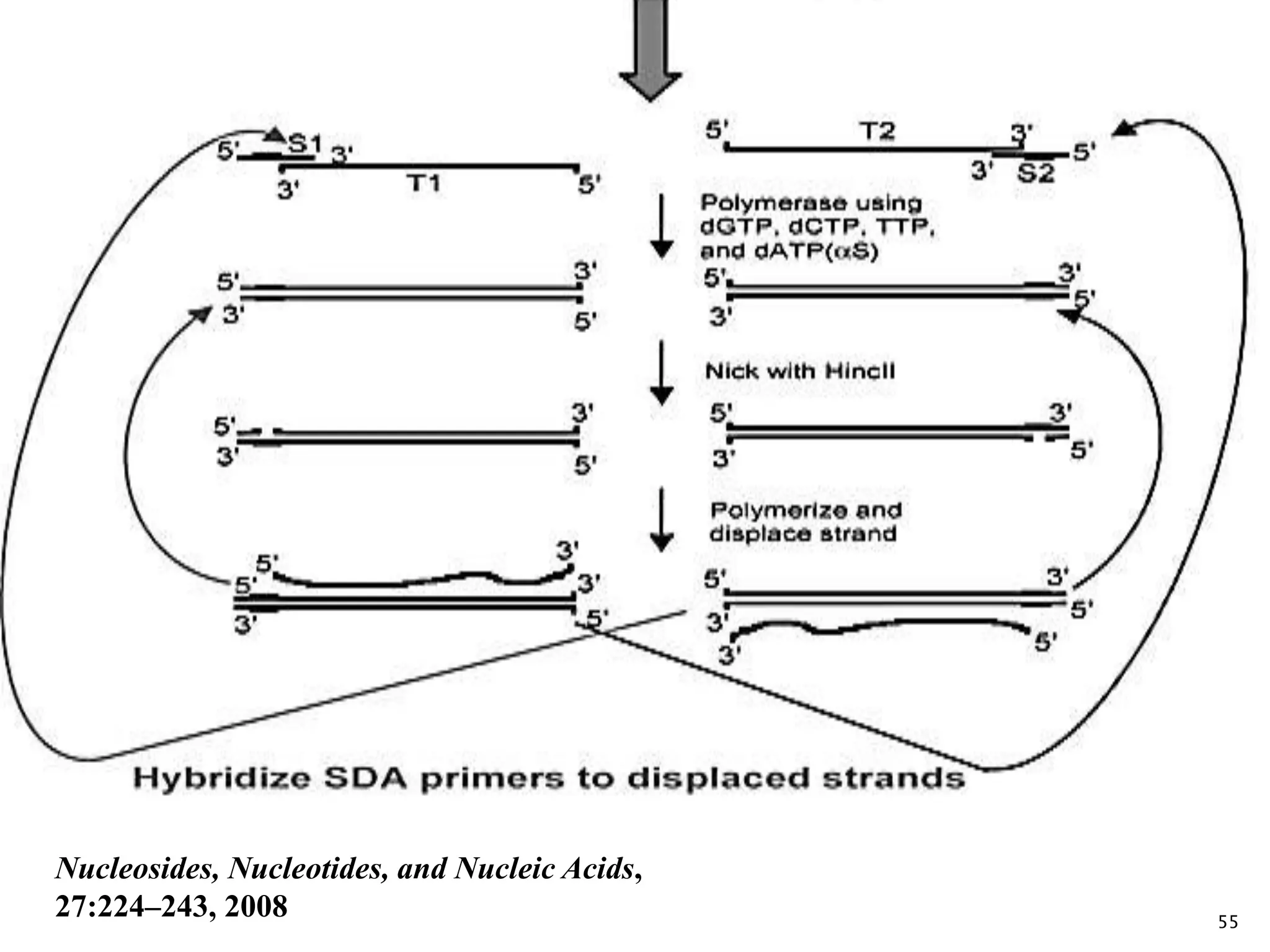
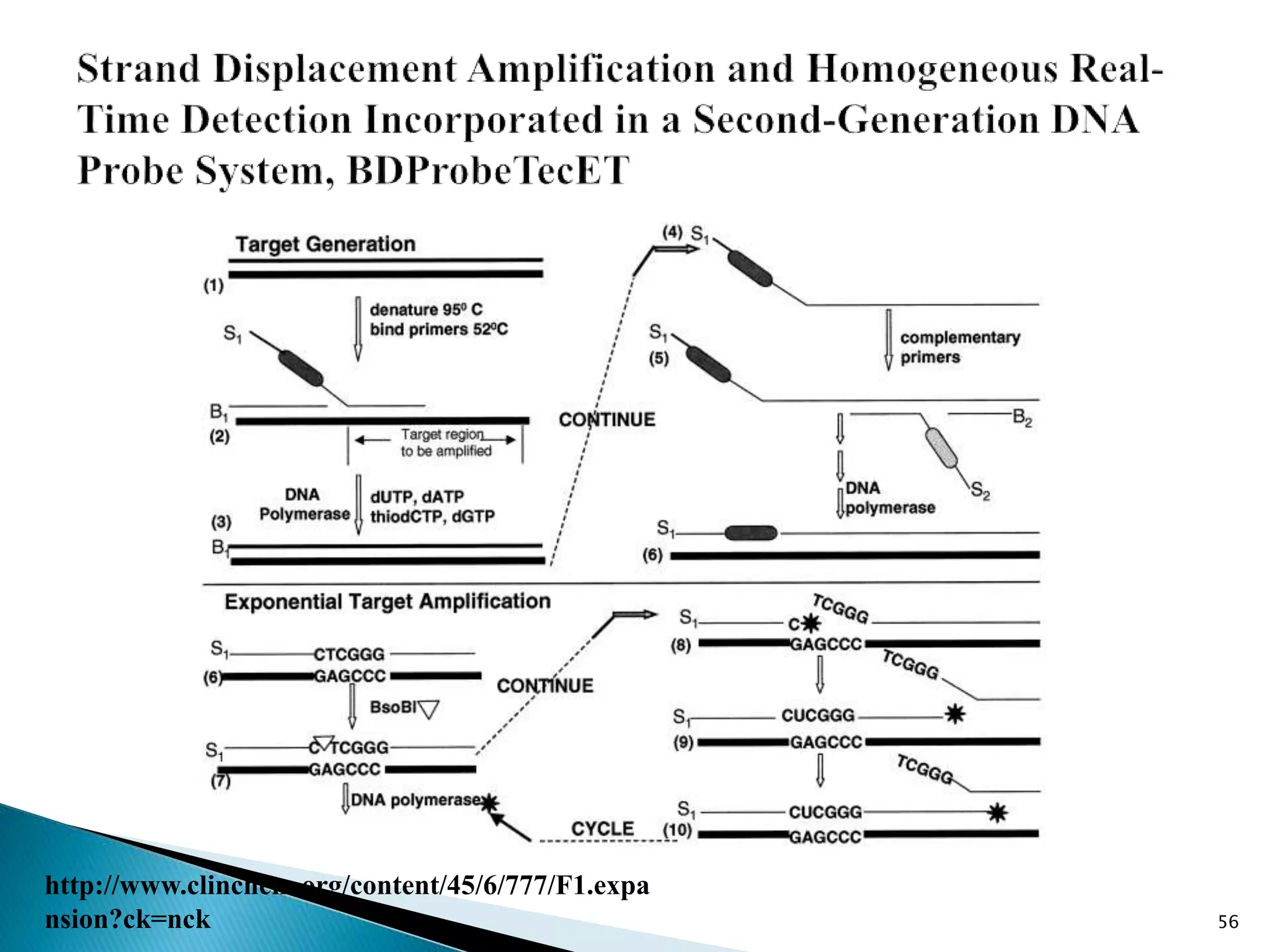
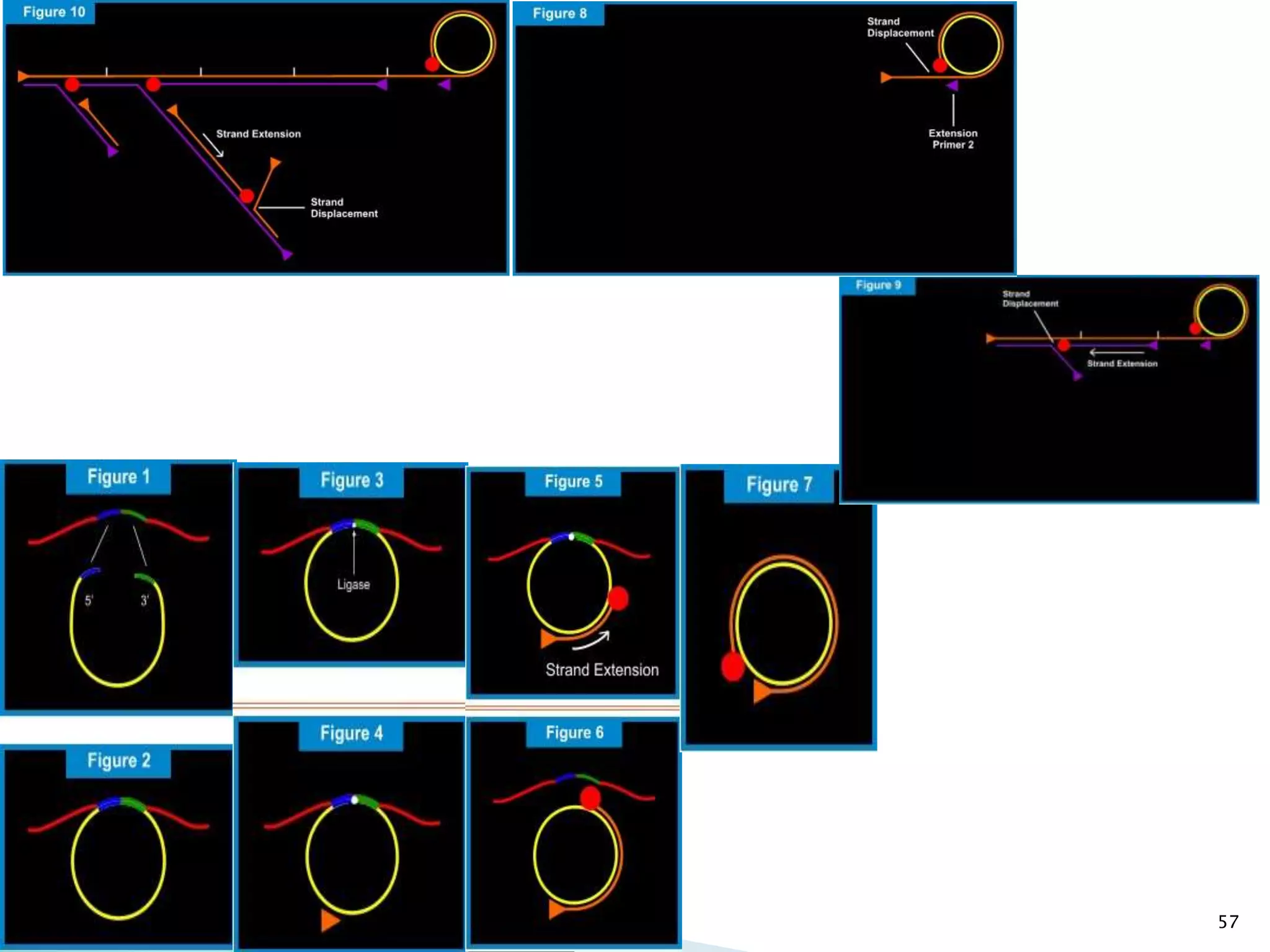
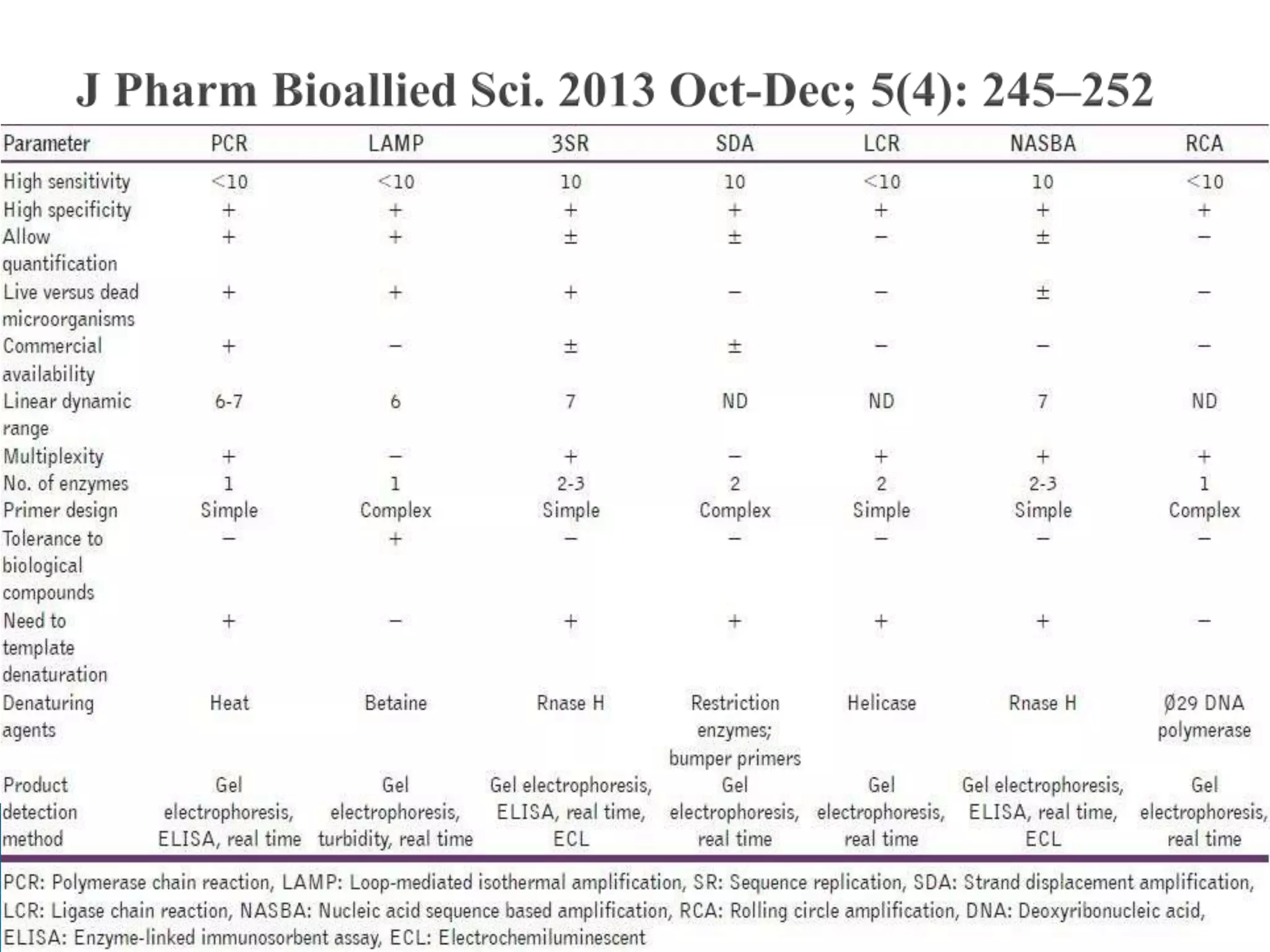
1. TMA, NASBA, SMART, SDA, RCA, LAMP, and others that amplify nucleic acids under isothermal conditions to overcome limitations of PCR like high costs and contamination risks.
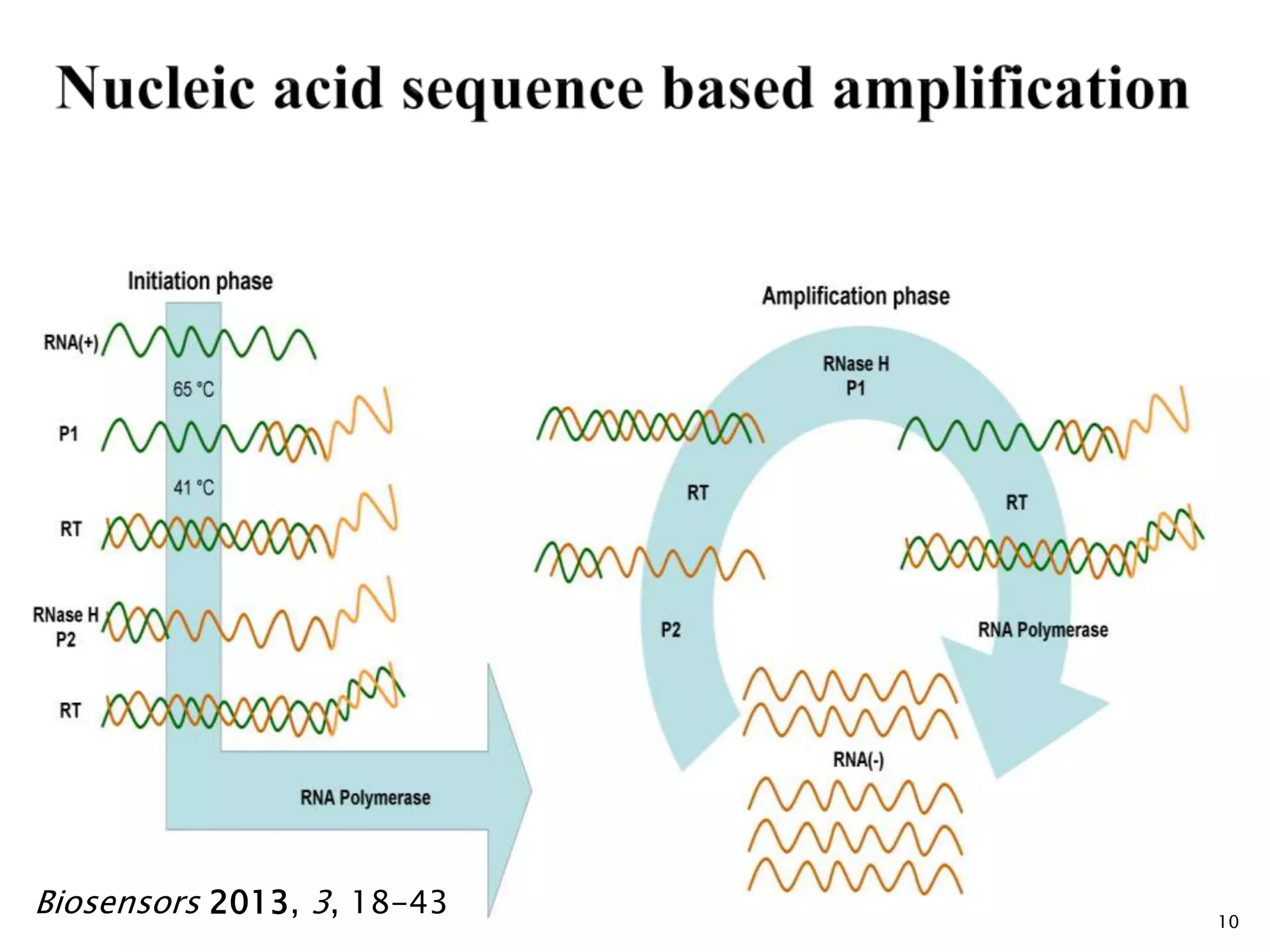
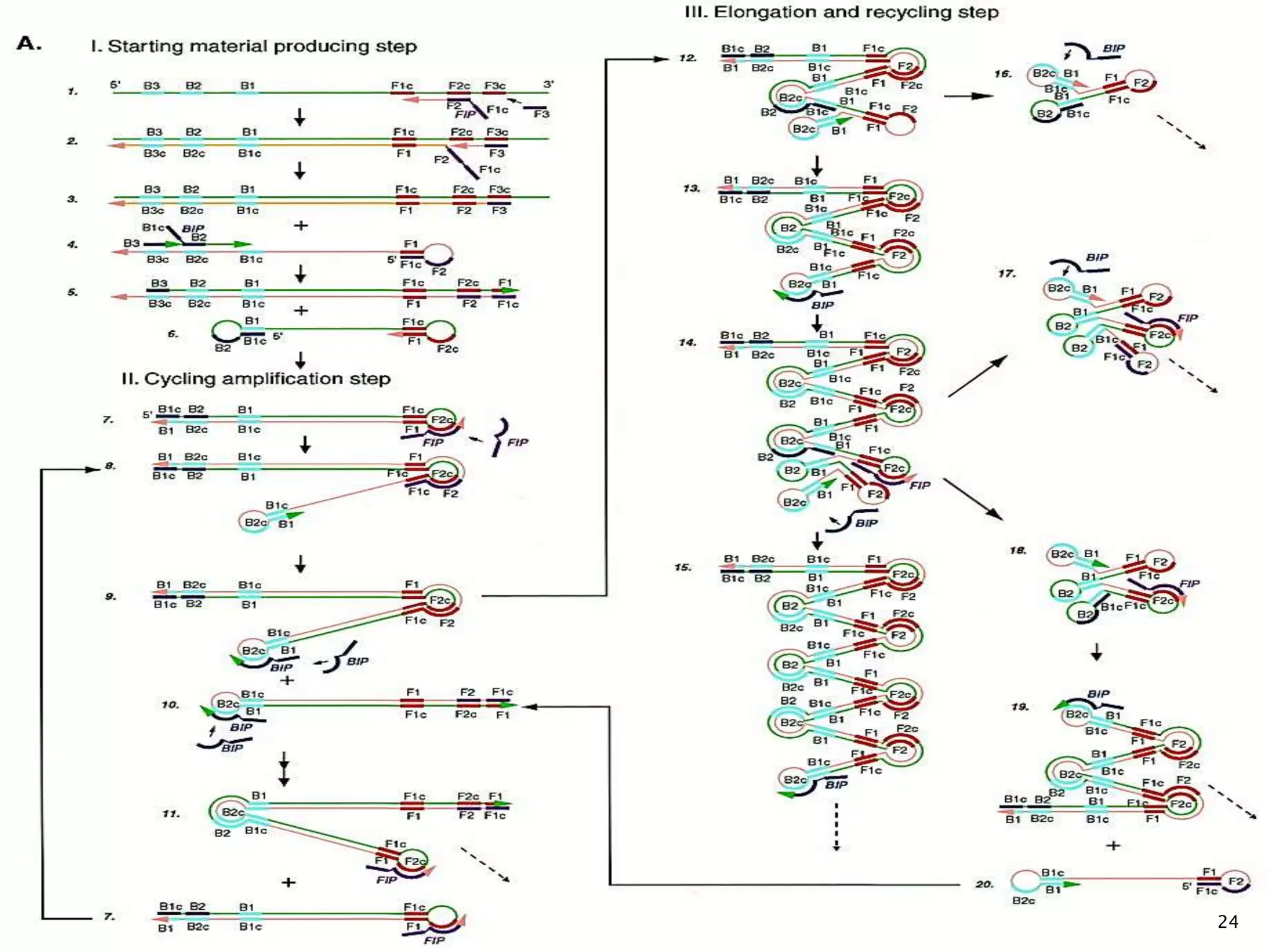
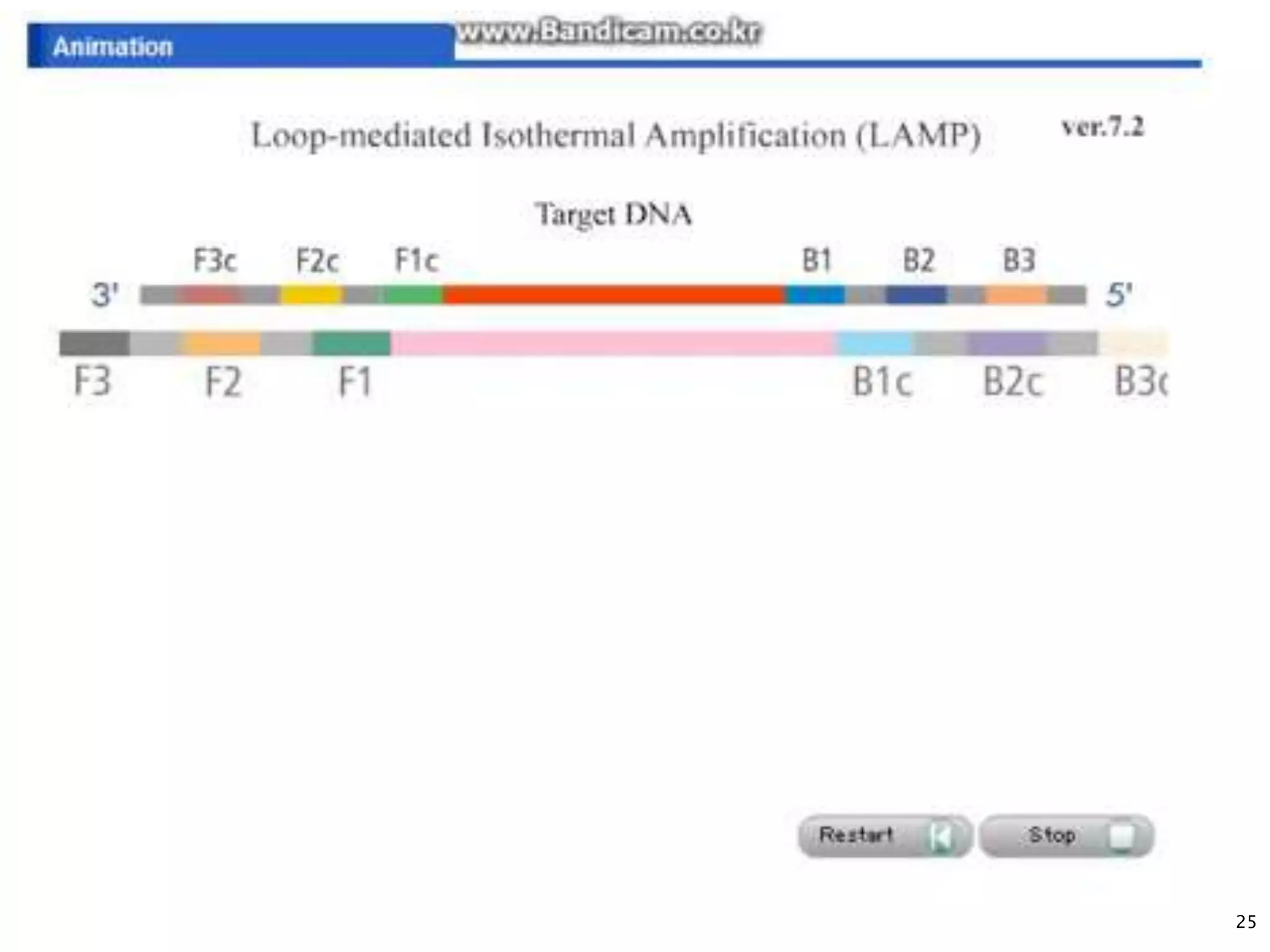
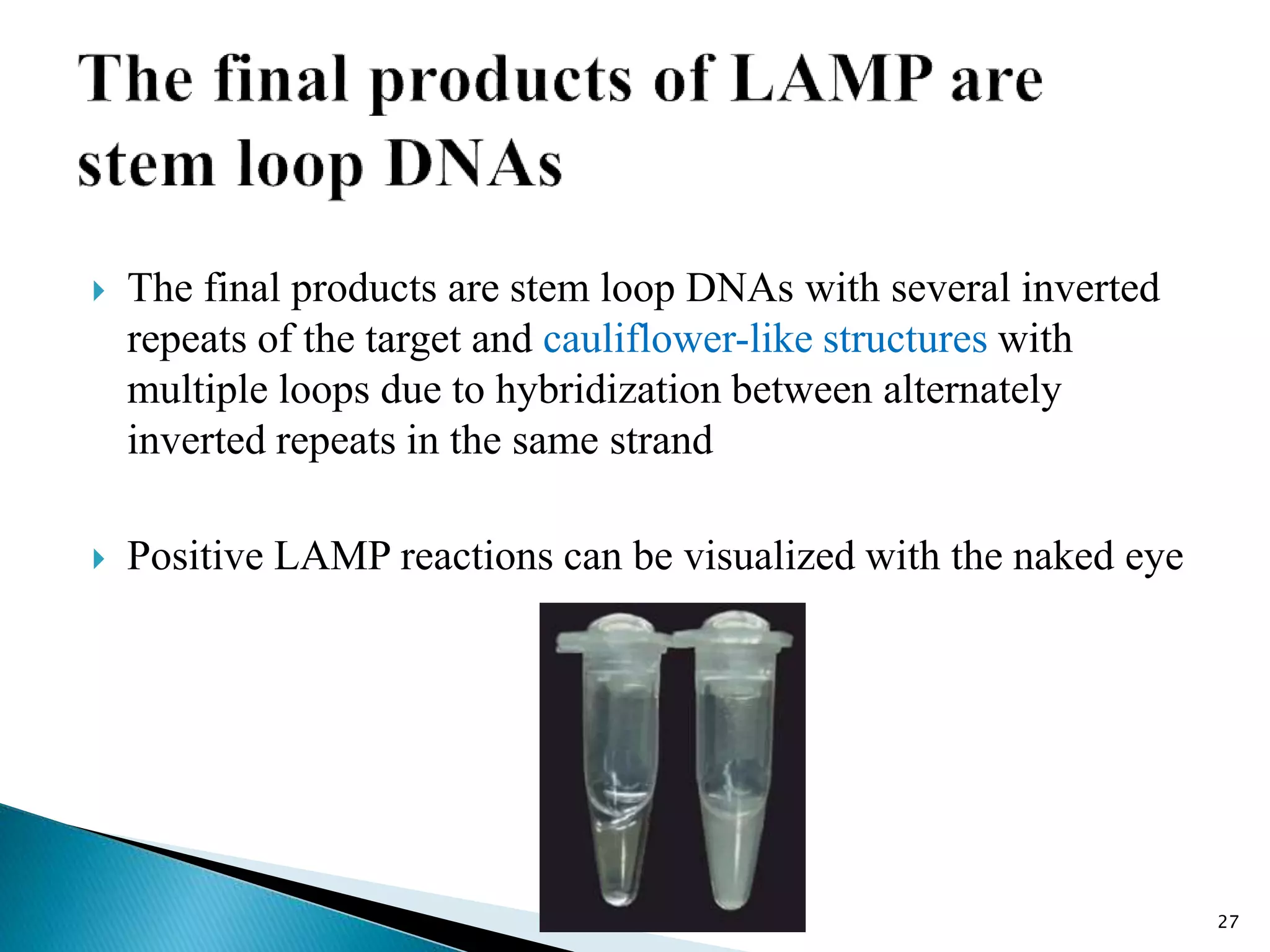
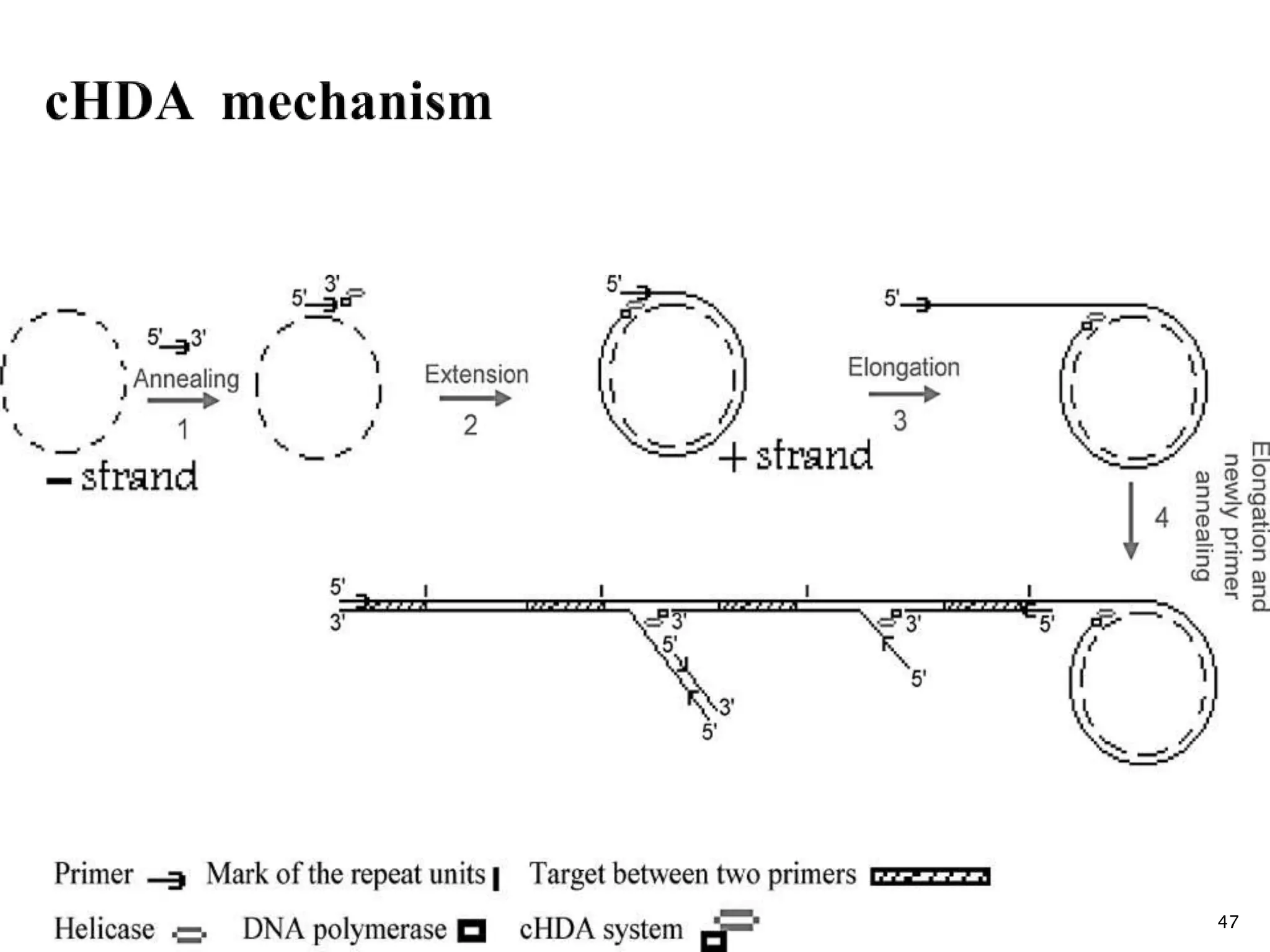
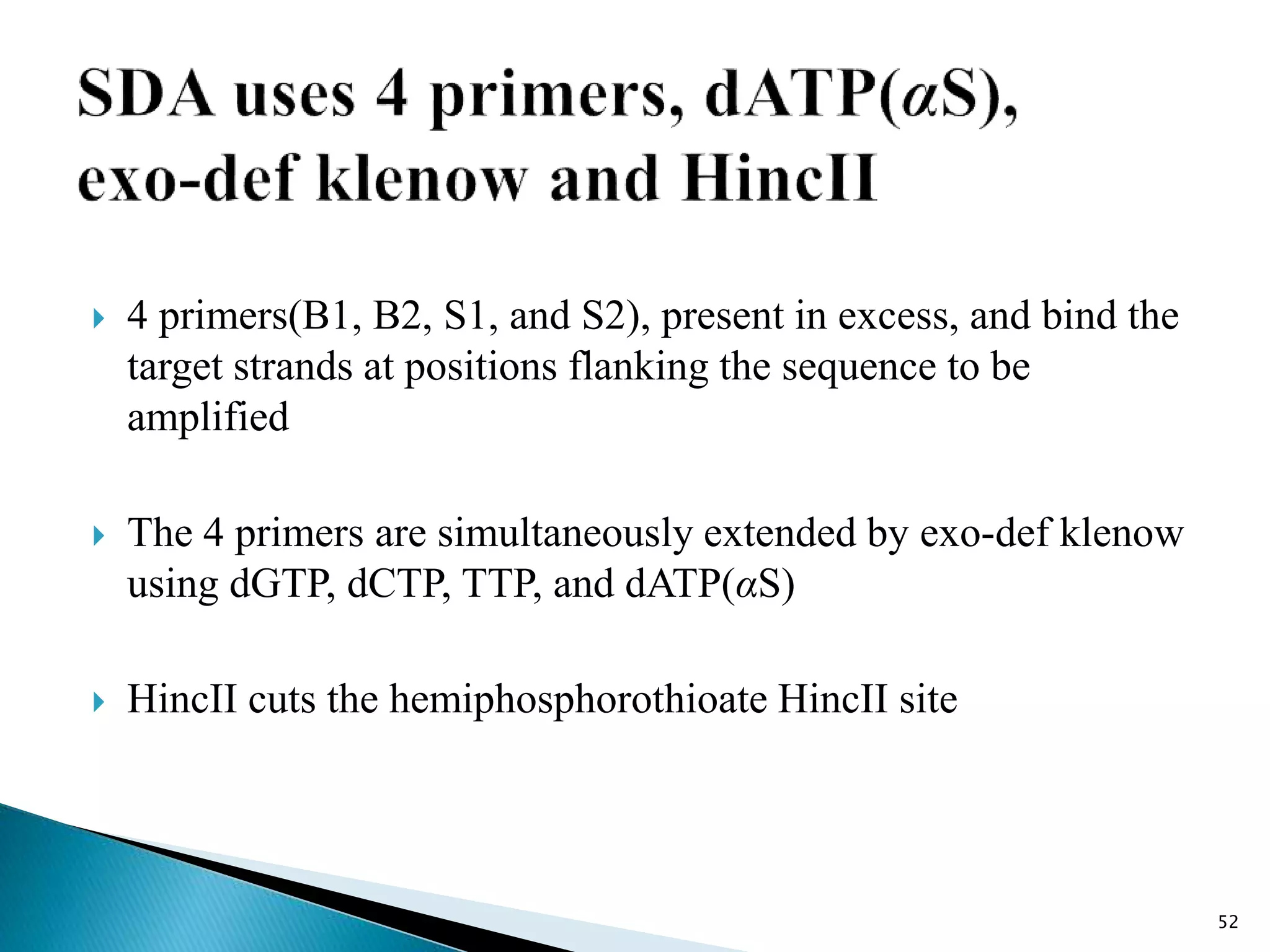
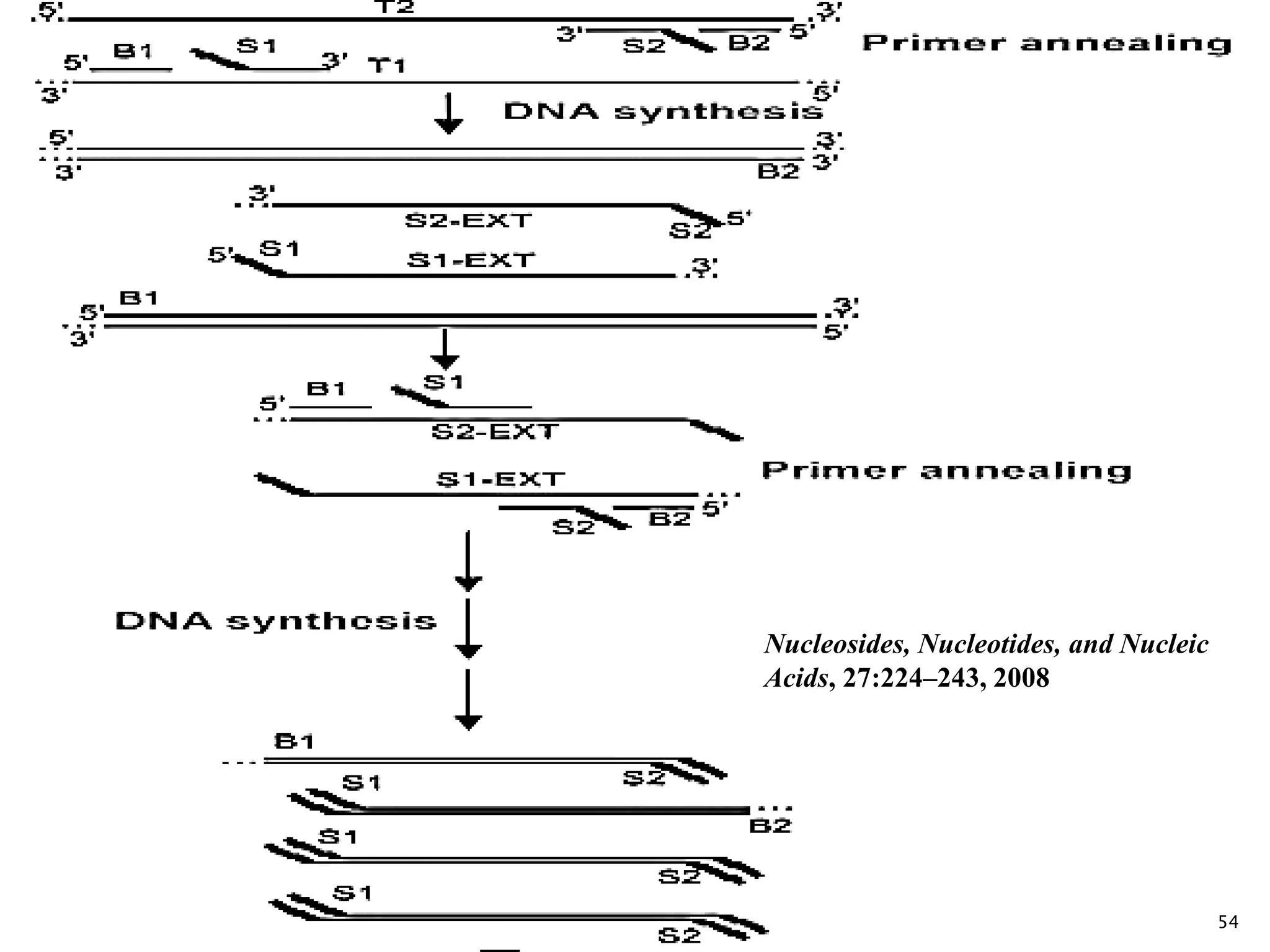
2. It describes several techniques in detail, explaining their mechanisms which involve primers, polymerases, and isothermal reactions to exponentially amplify target sequences.
3. The techniques have applications in research, medicine, forensics and more due to their simplicity, lower costs, and ability to be used at point-of-care without specialized equipment.