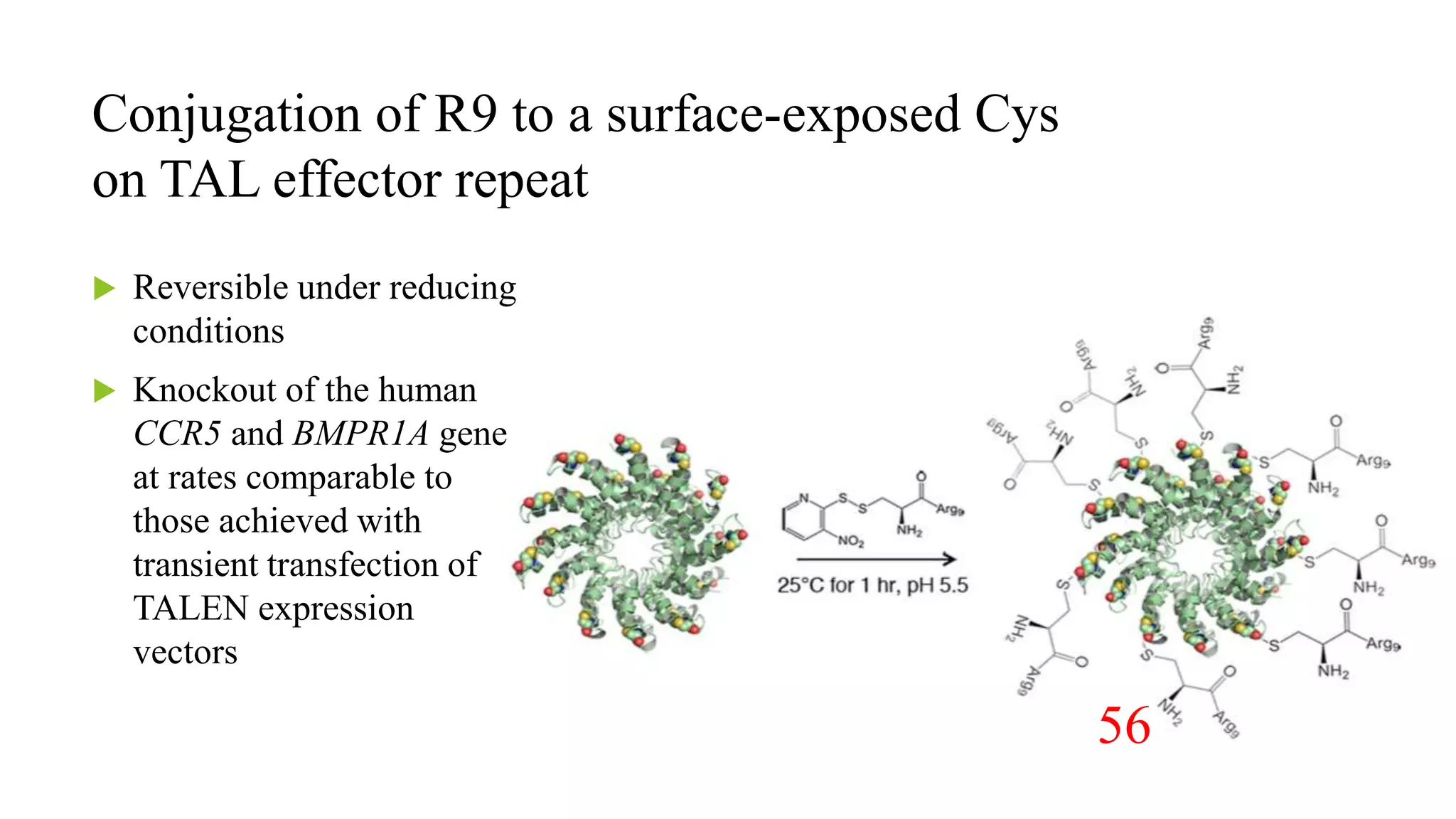
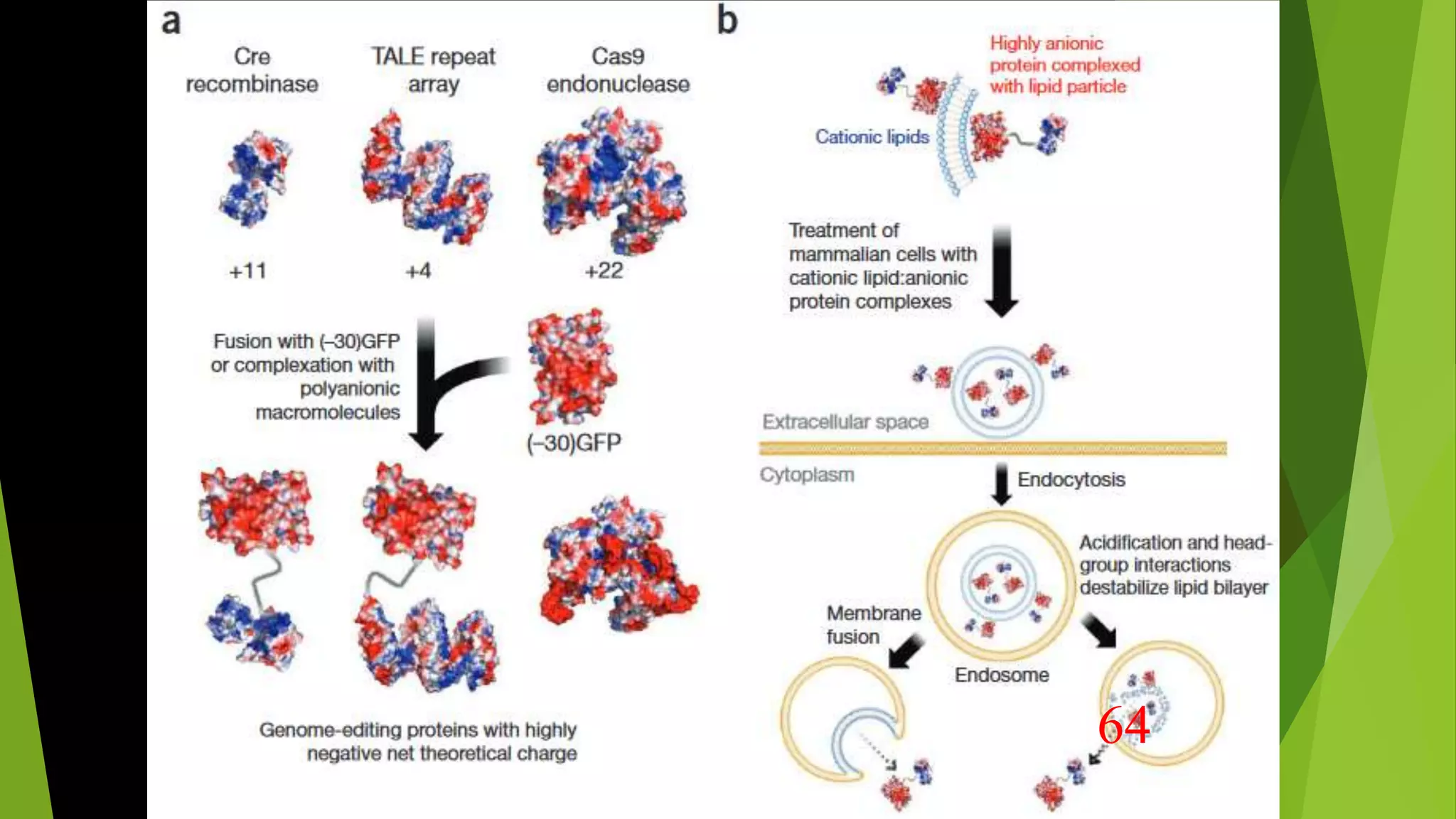
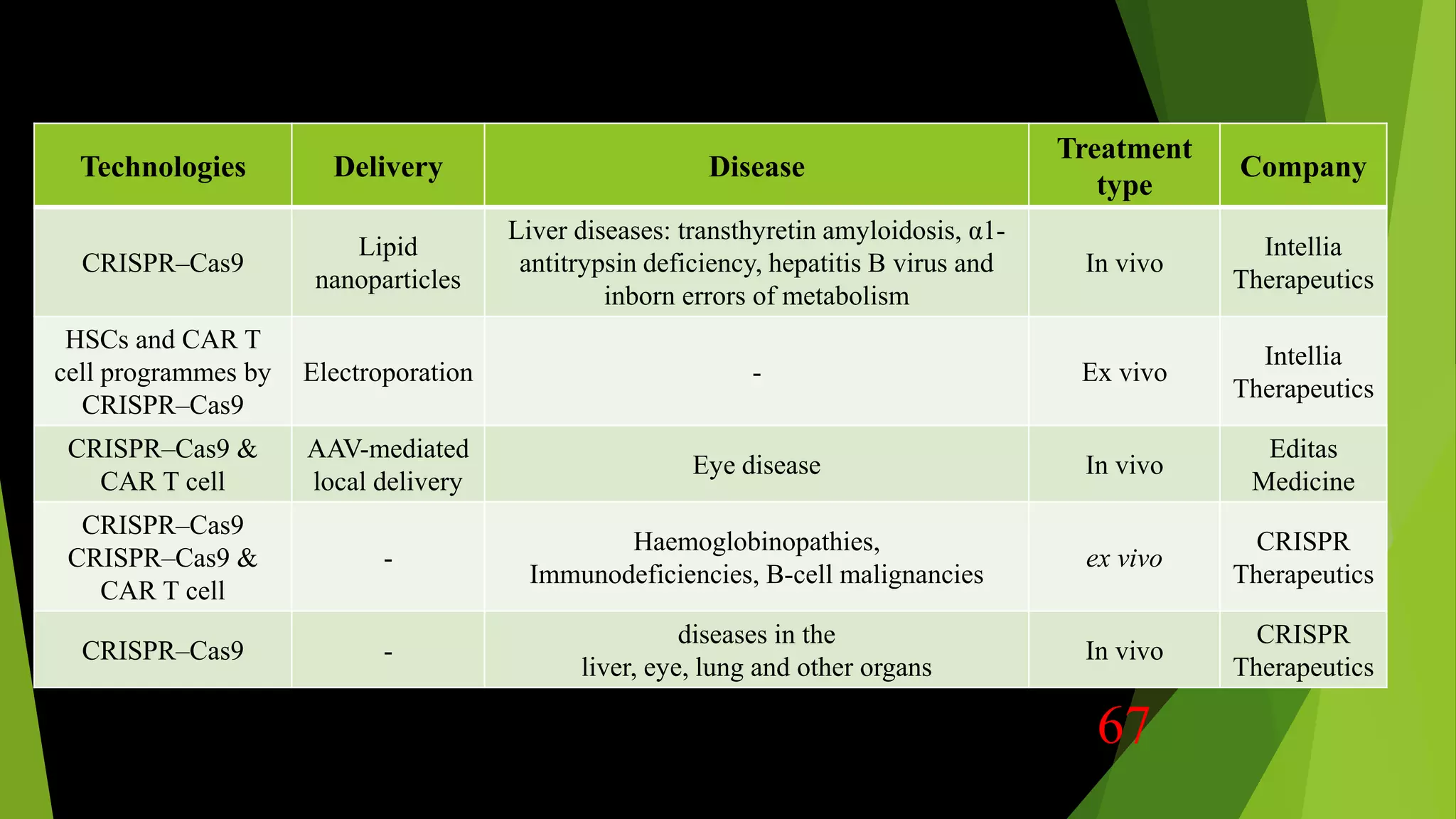
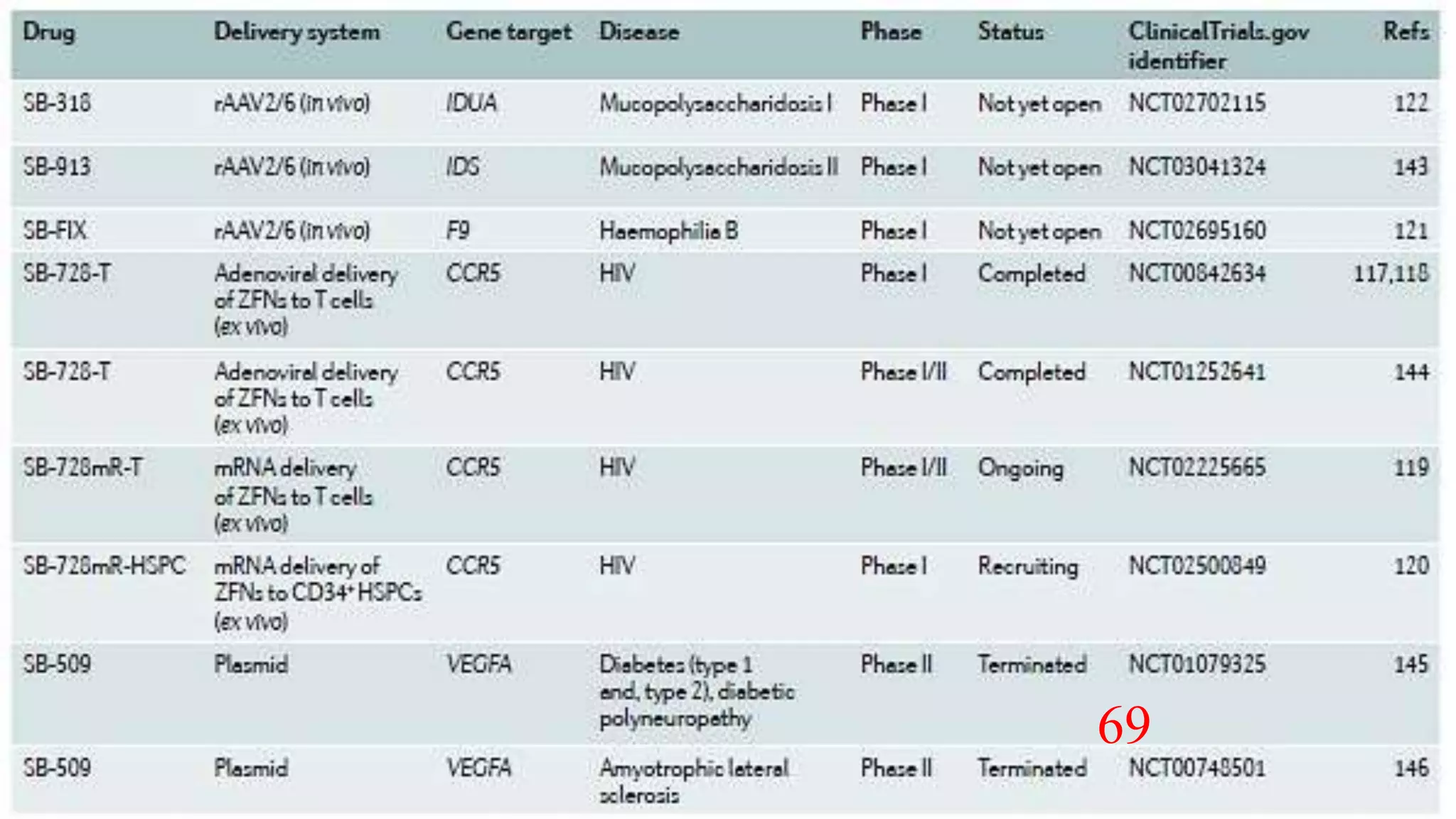
The document discusses advancements in genome editing technologies, highlighting the need for effective therapies for genetic diseases and the challenges associated with delivering these therapies in vivo and ex vivo. It covers various approaches such as viral and non-viral delivery methods, RNA modification therapies, and the application of CRISPR/Cas9 technology for treating conditions like hemophilia and hereditary diseases. Additionally, it emphasizes the importance of precise gene editing and the limitations related to off-target effects and delivery mechanisms.















































![The most widely investigated physical transfection
method is electroporation
Electroporation of Cas9 and two different sgRNA-encoding plasmids
significant gene deletion in HSPCs and in primary T cells
Electroporation of [Cas9 + in vitro transcribed sgRNA ] RNP
Higher efficiency genome editing
Decreased off-target effects
Less cellular toxicity
Non-Viral ---- In vitro & ex vivo
49](https://image.slidesharecdn.com/genomeeditingdelivery-200809034059/75/Genome-editing-delivery-systems-49-2048.jpg)