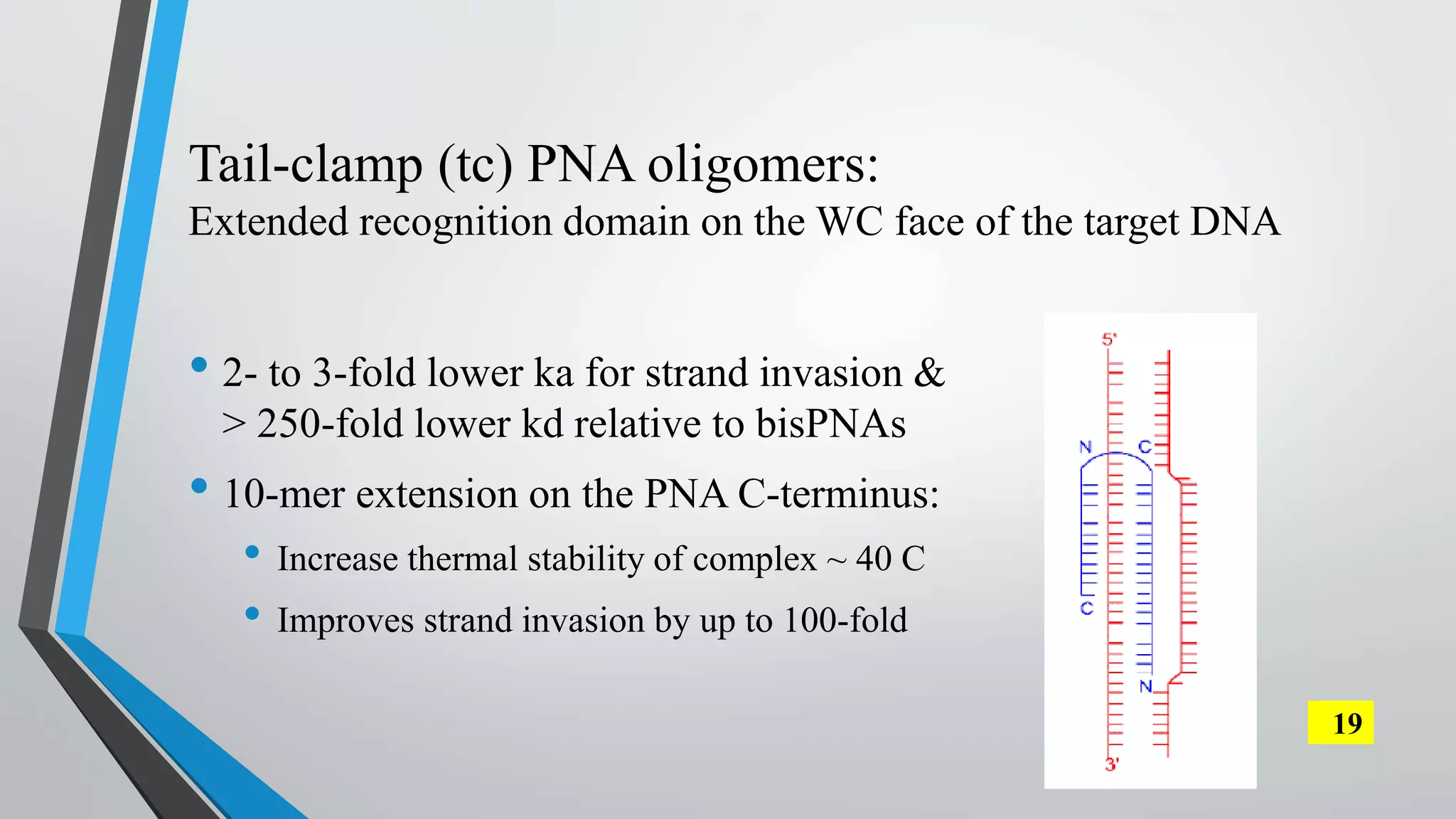
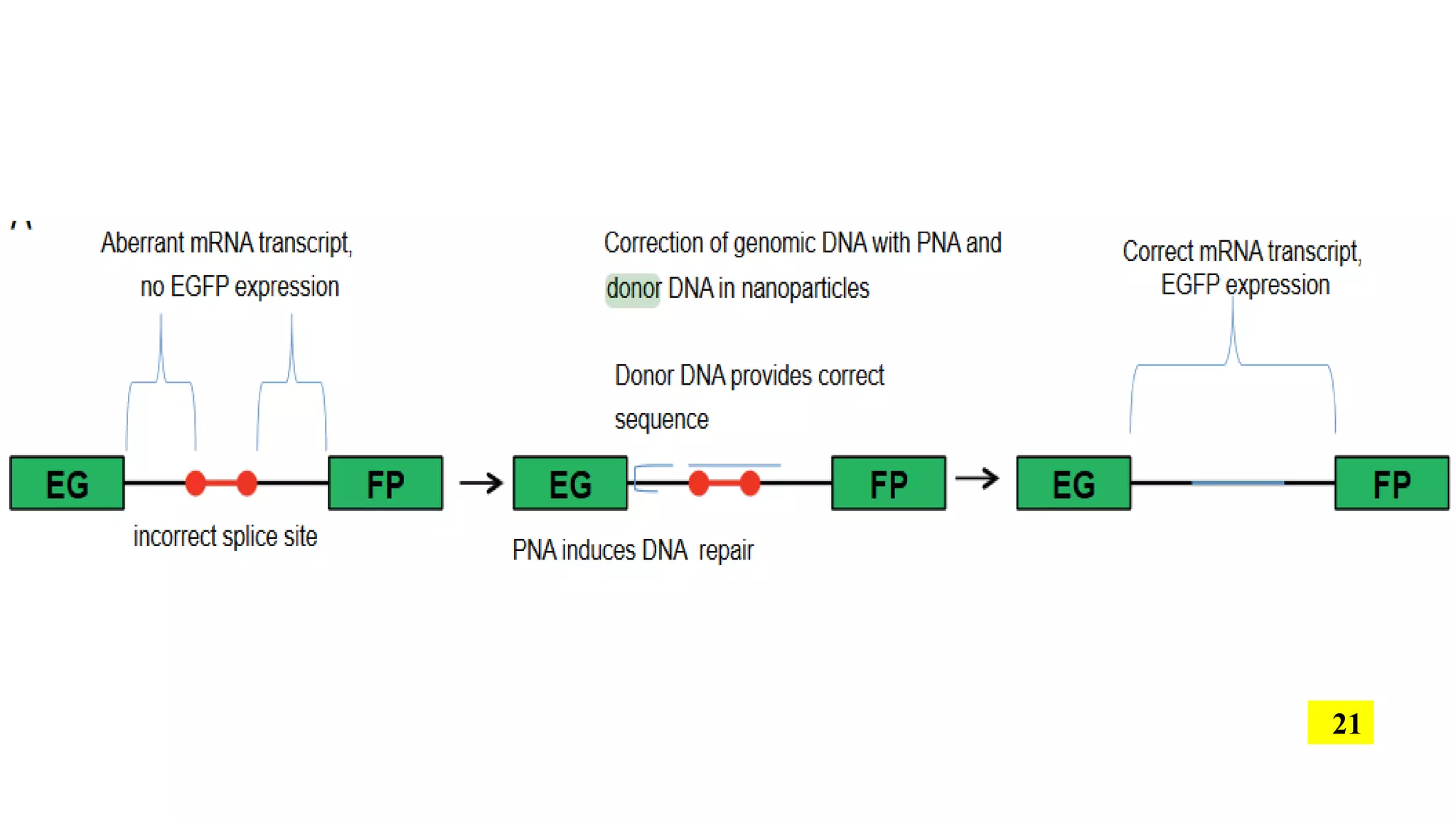
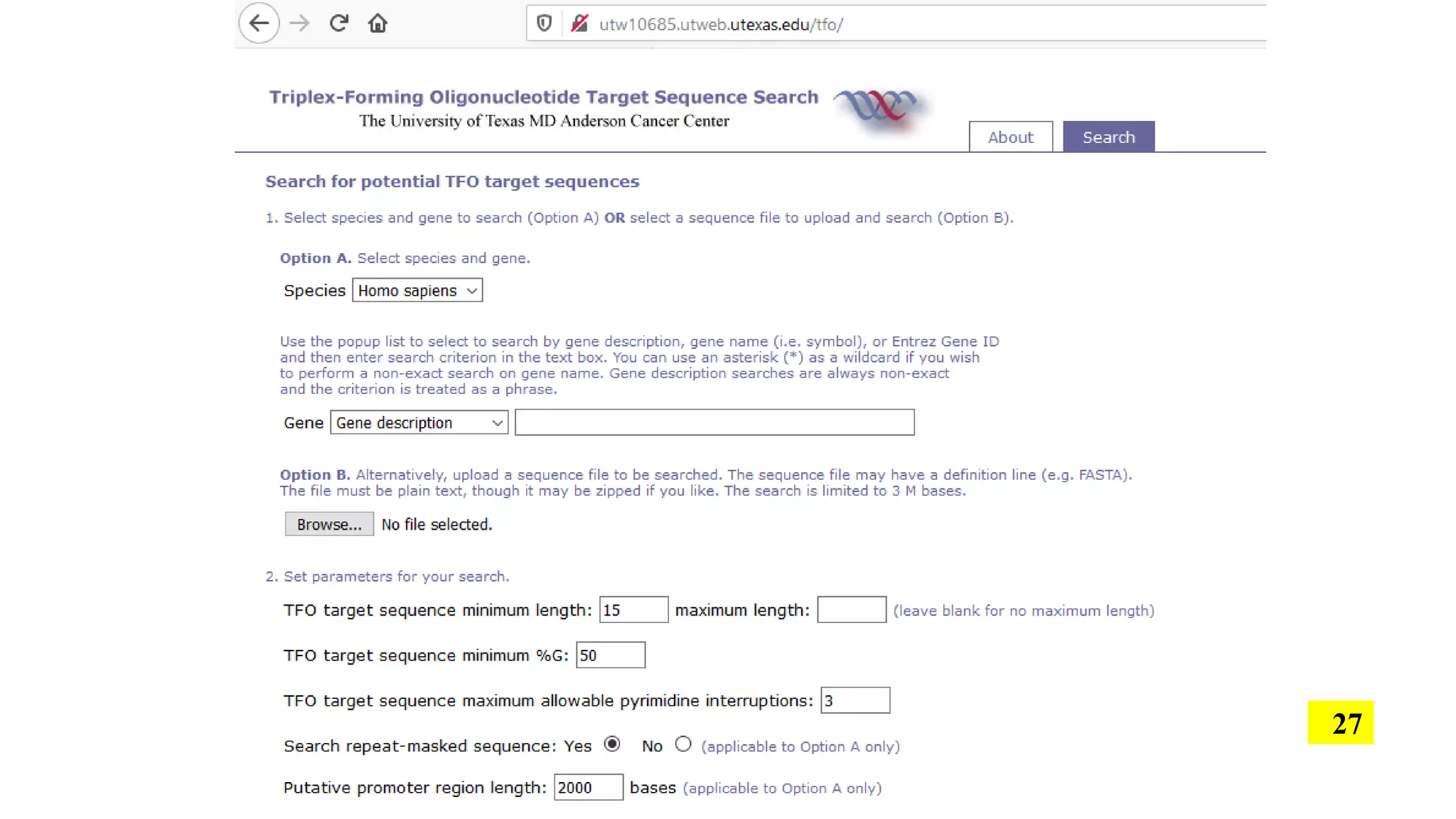
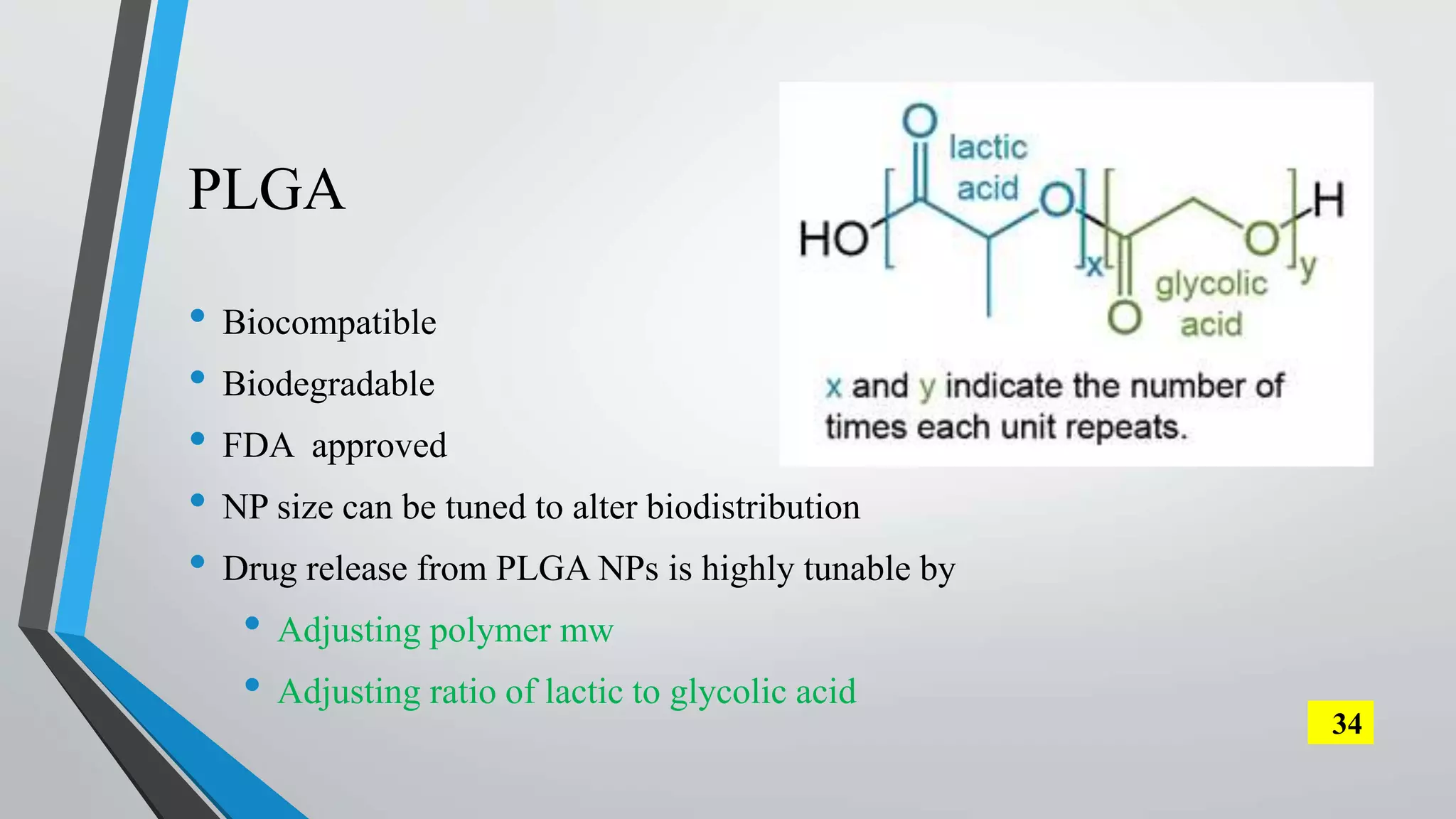
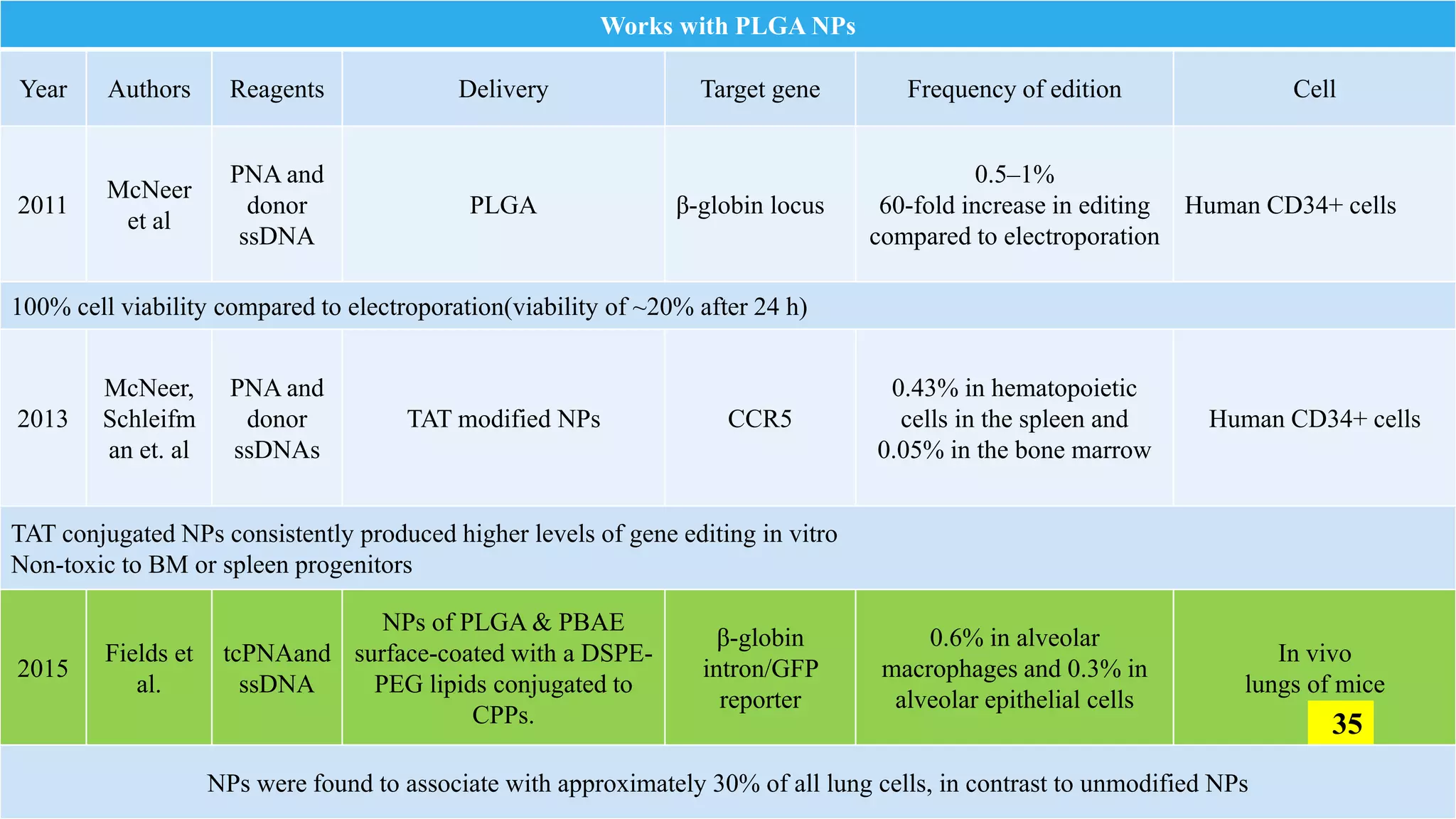
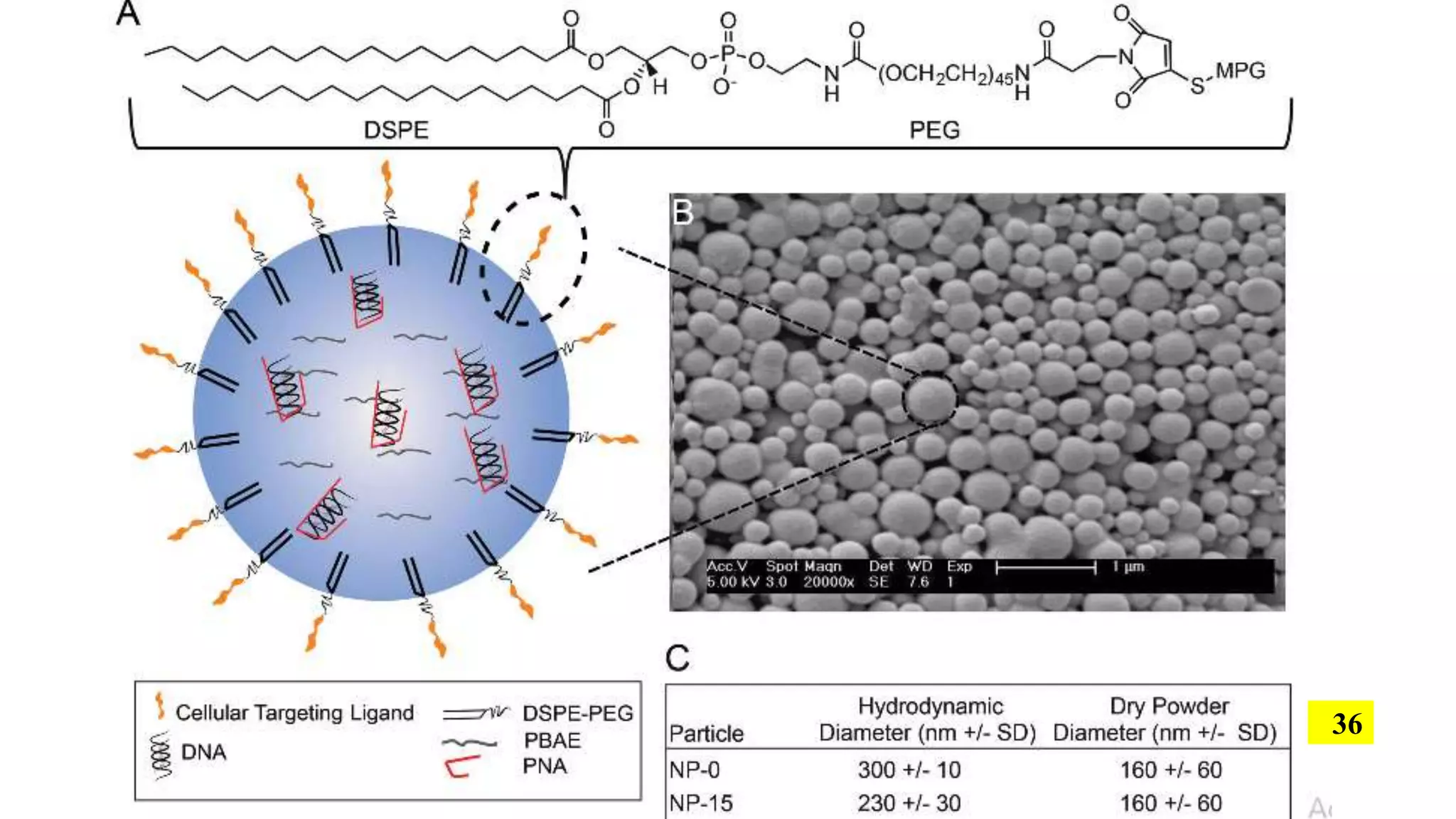
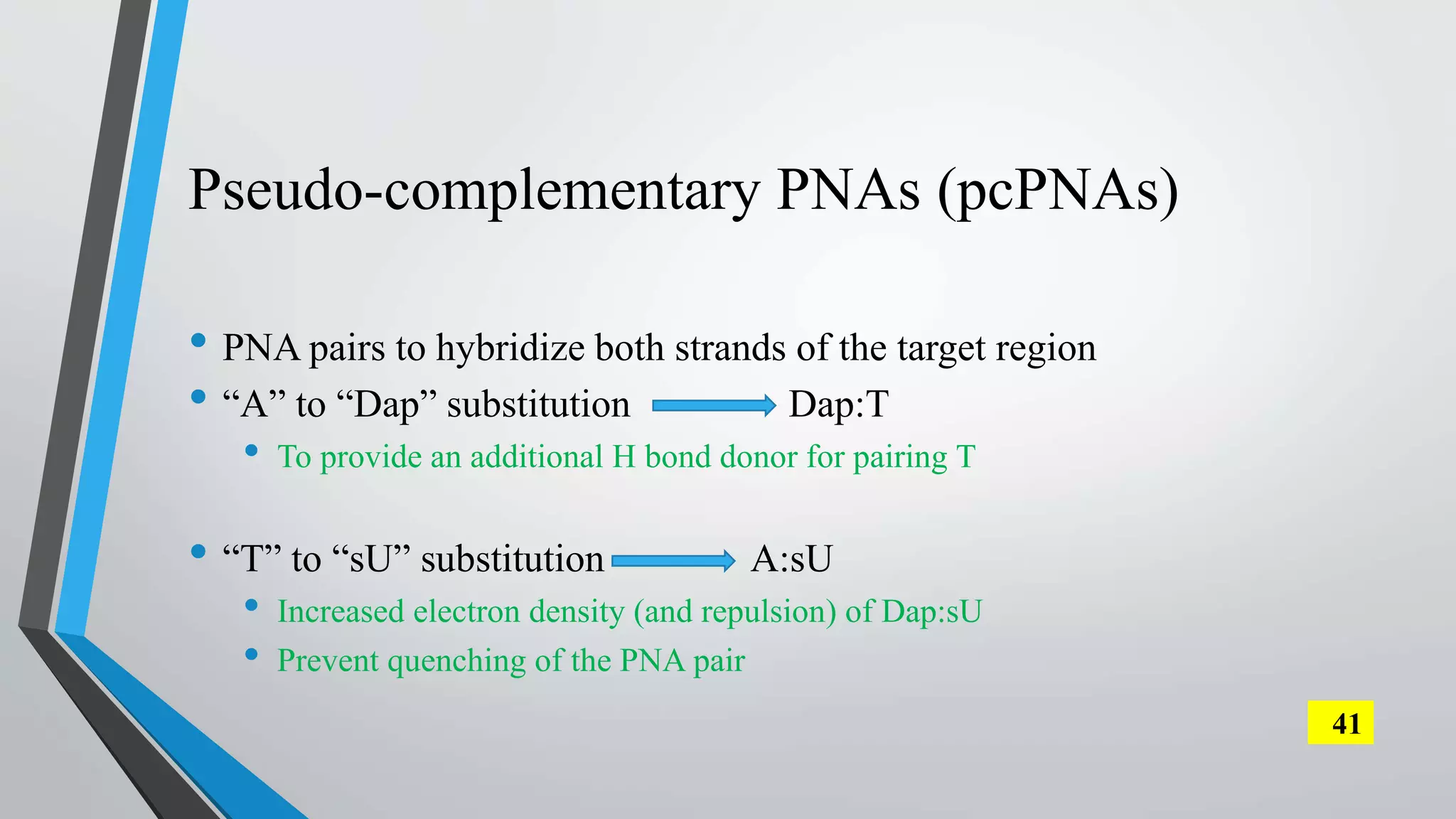
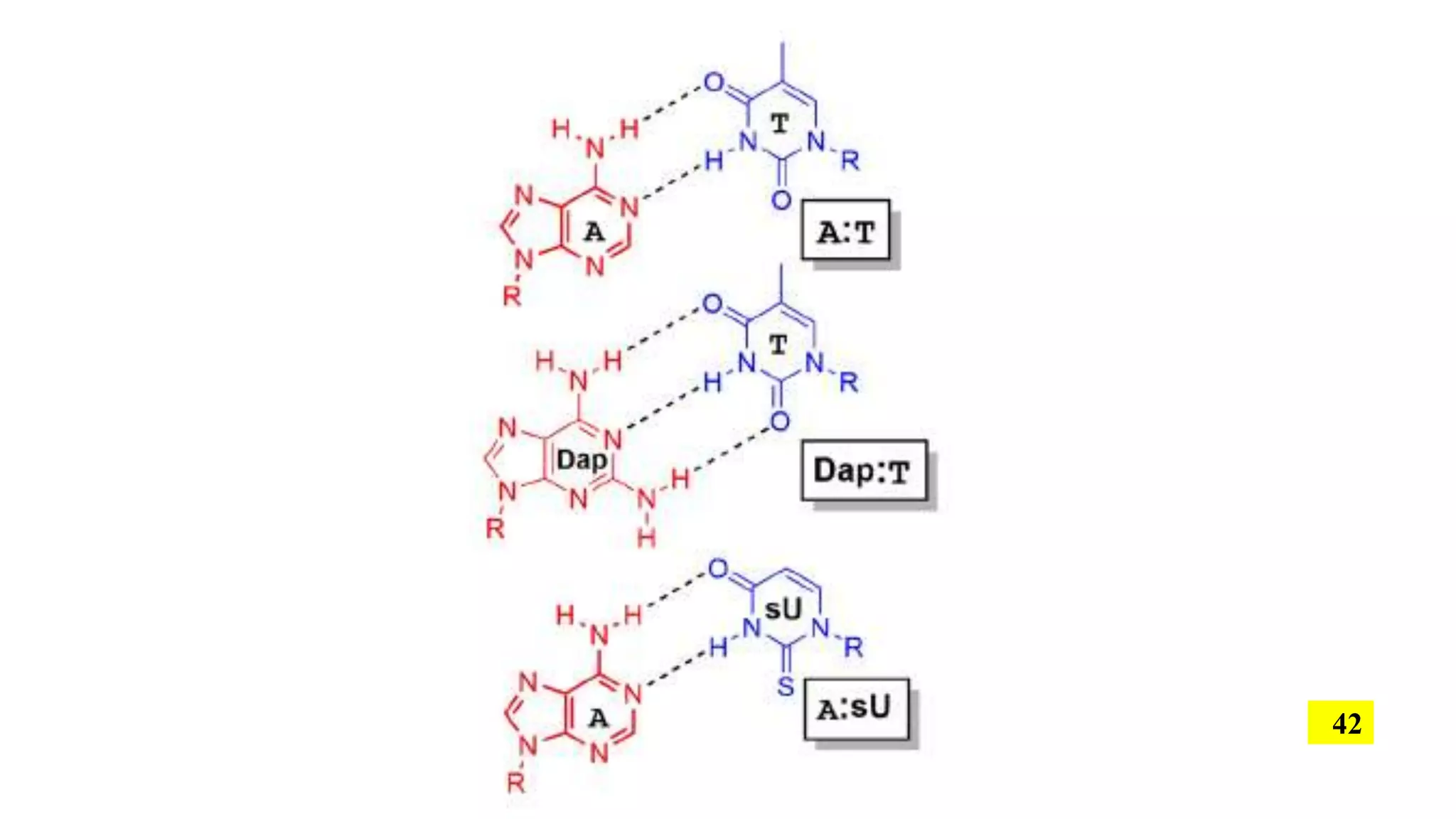
The document discusses the development and applications of peptide nucleic acids (PNA) in gene editing, outlining their structural advantages over traditional DNA. It highlights the successful utilization of various PNA technologies, including triplex-forming oligonucleotides and their delivery methods, for targeted genetic modifications across different cellular contexts. Additionally, it addresses the mechanisms of PNA-mediated gene editing and the challenges faced in achieving efficient cellular uptake and editing frequencies.