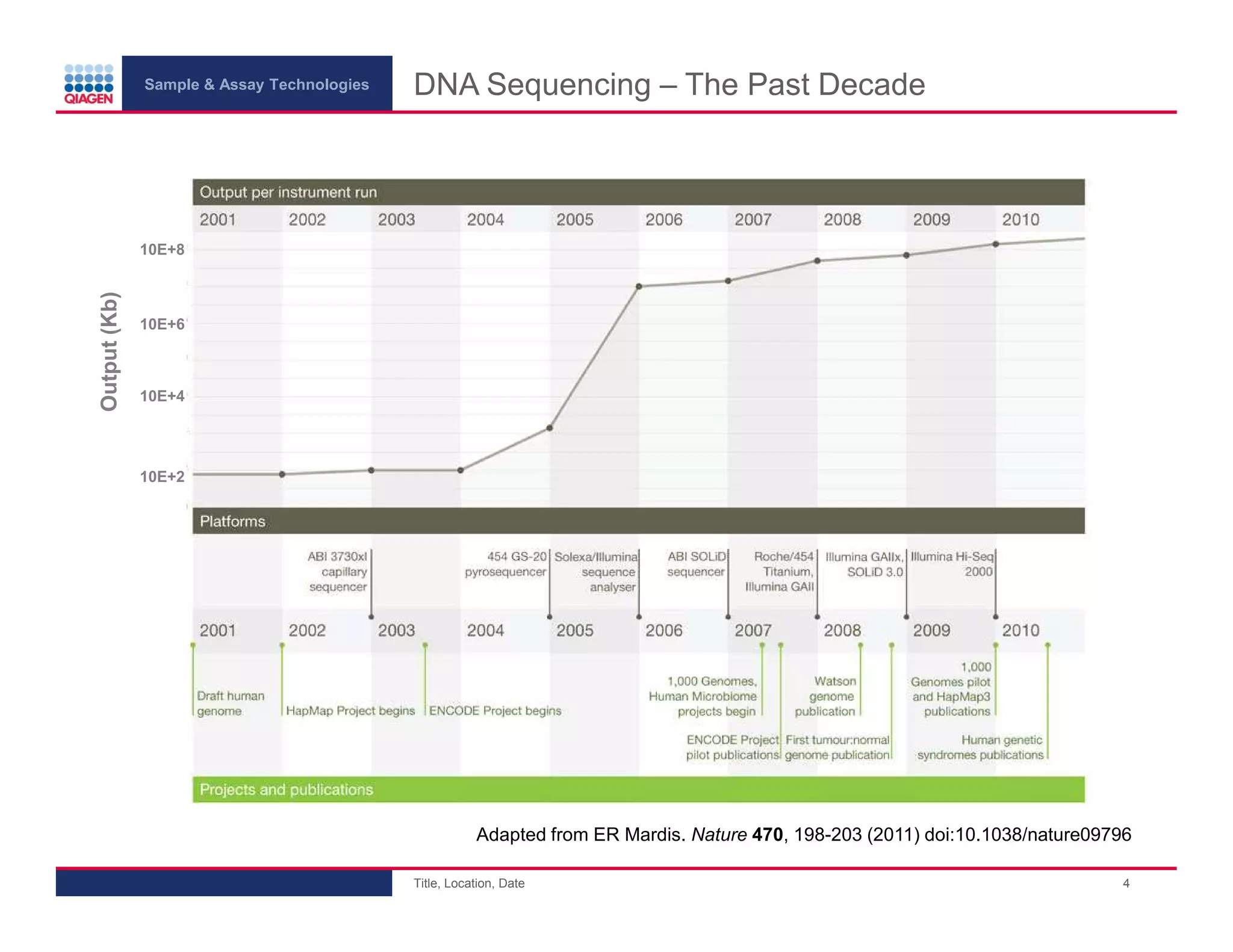
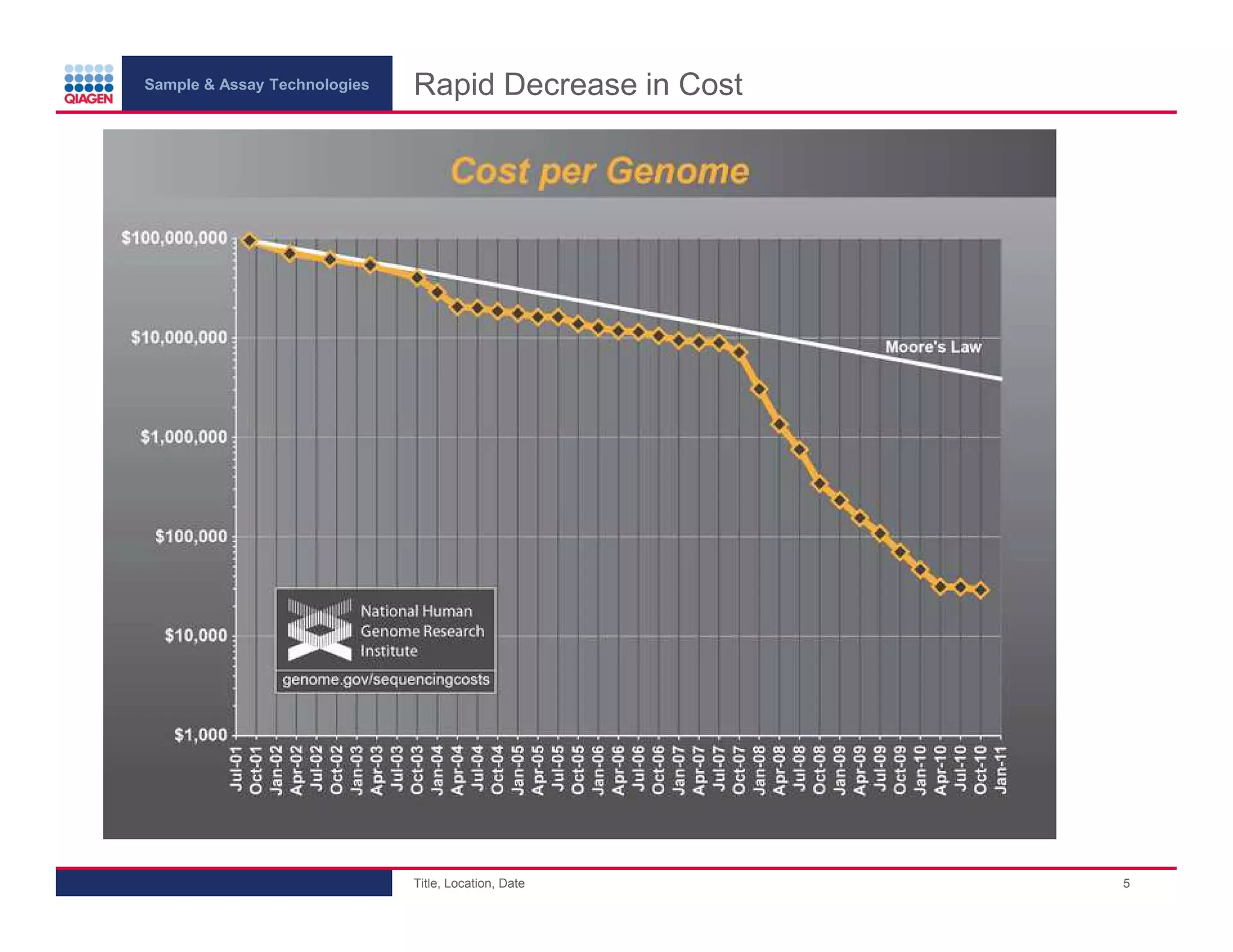
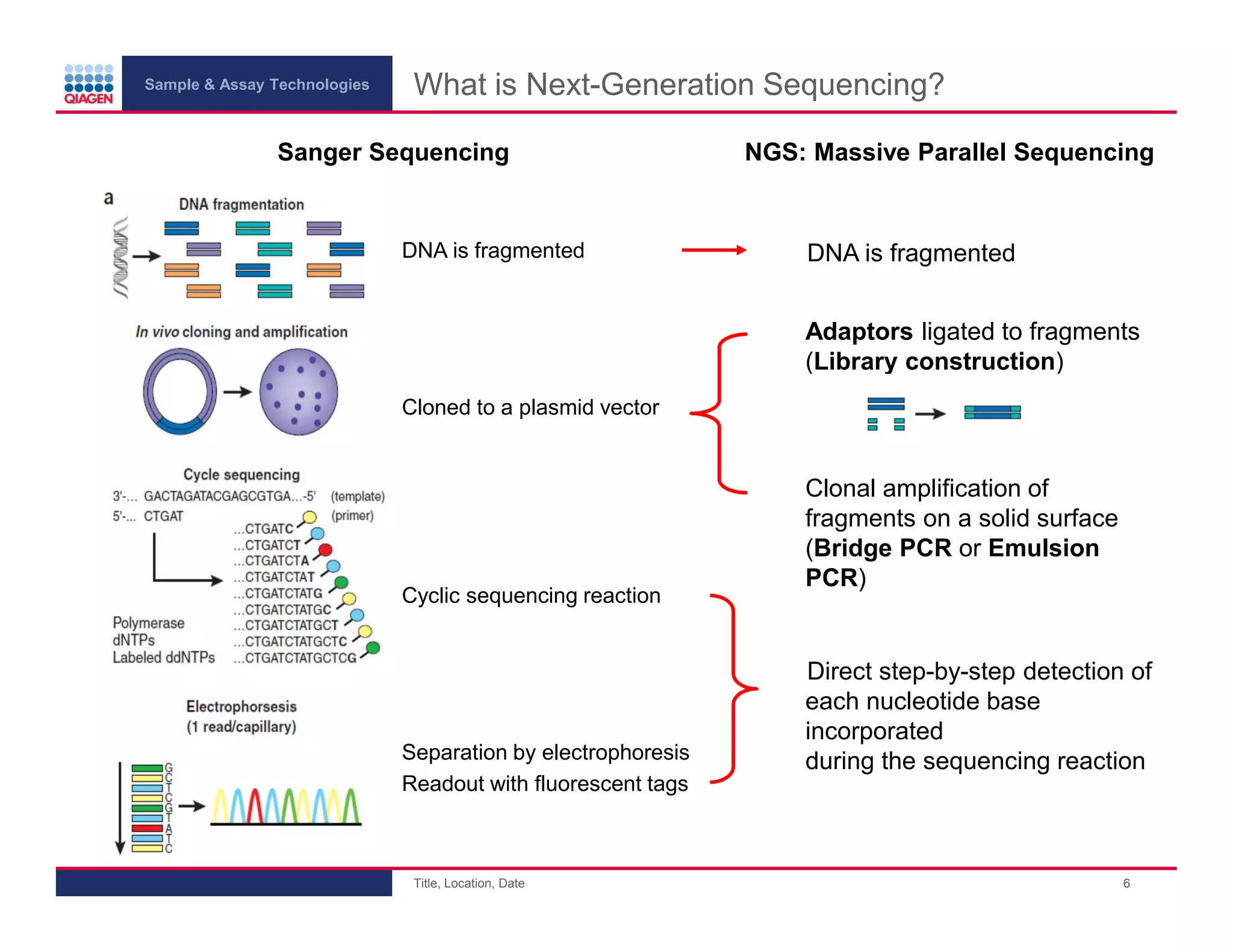
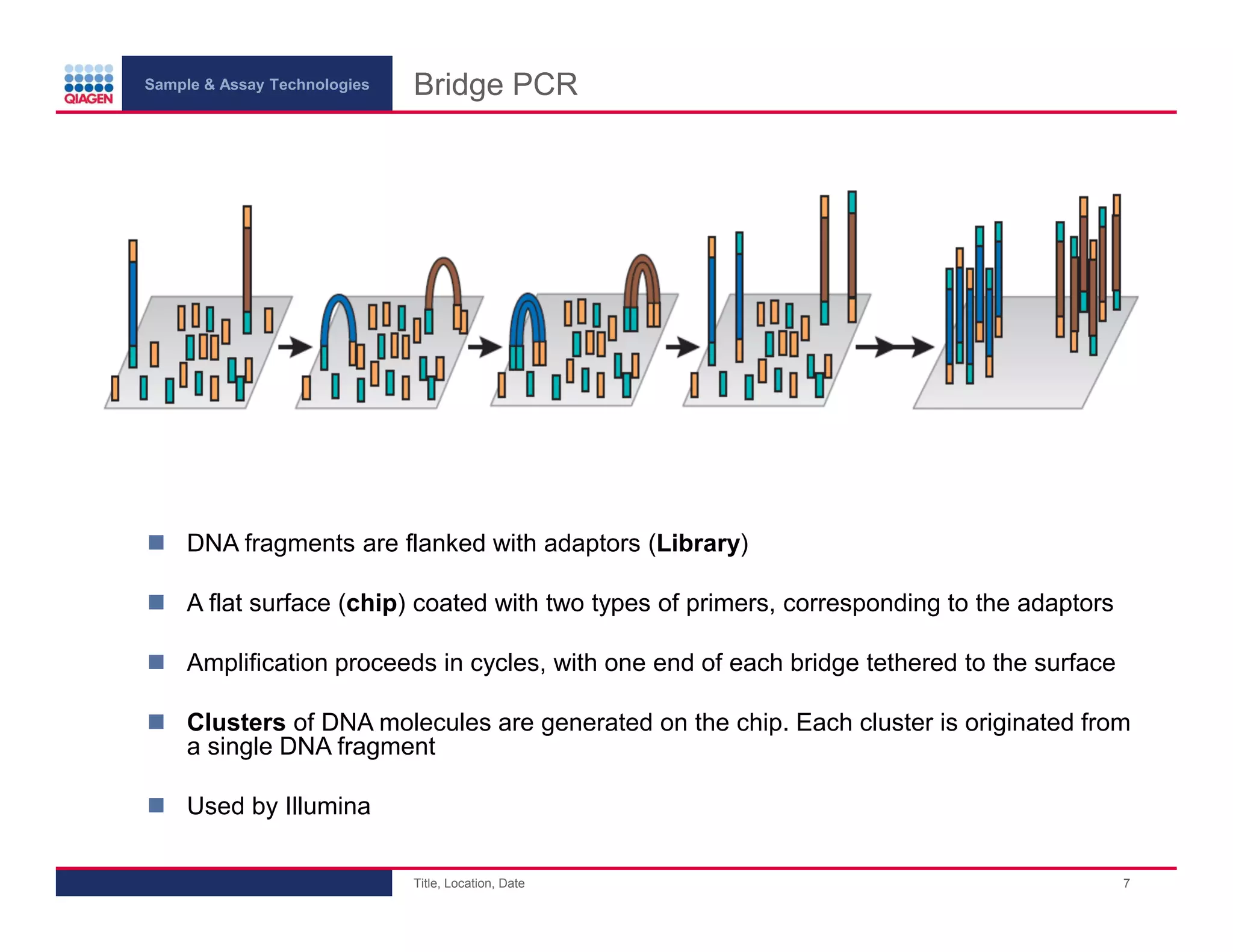
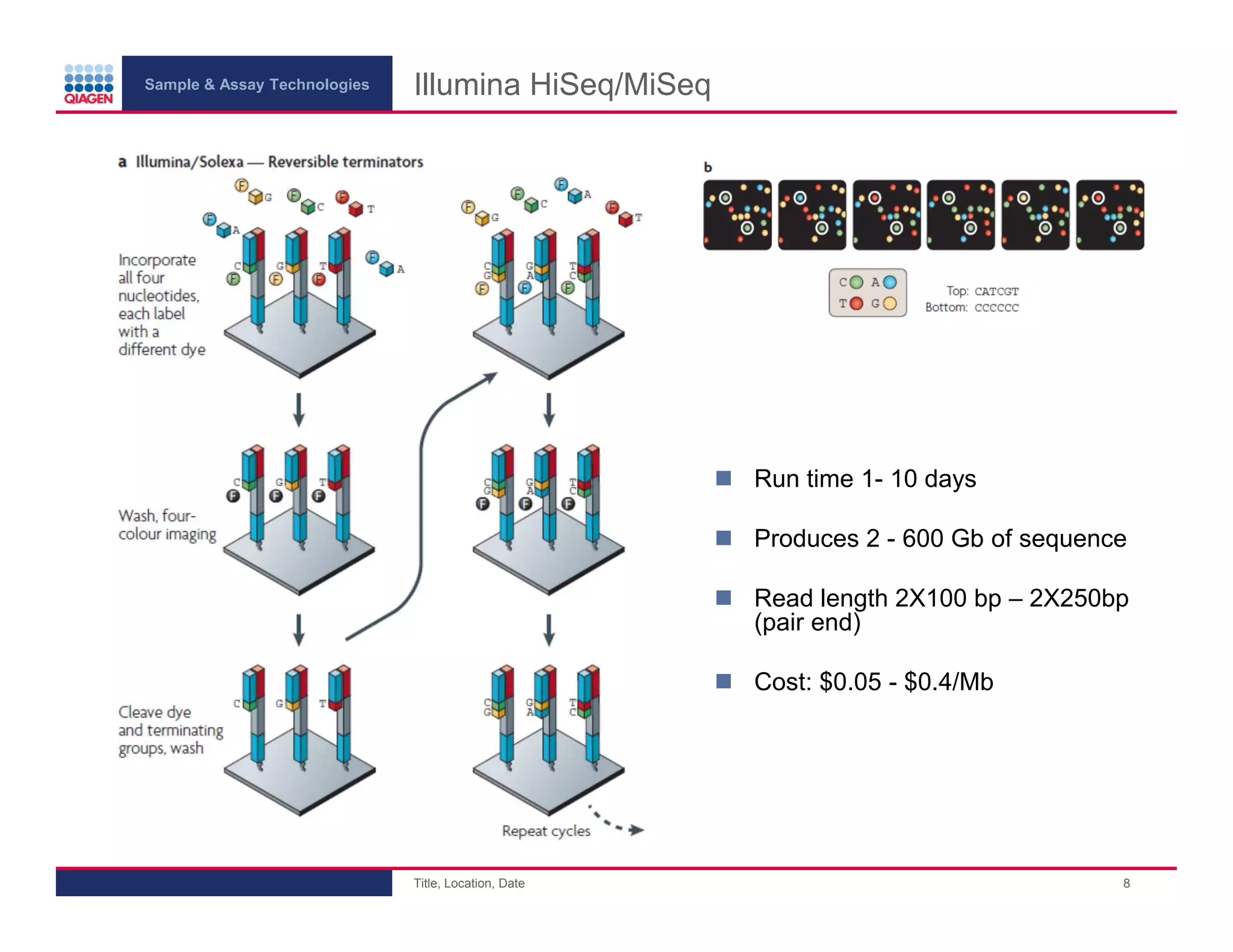
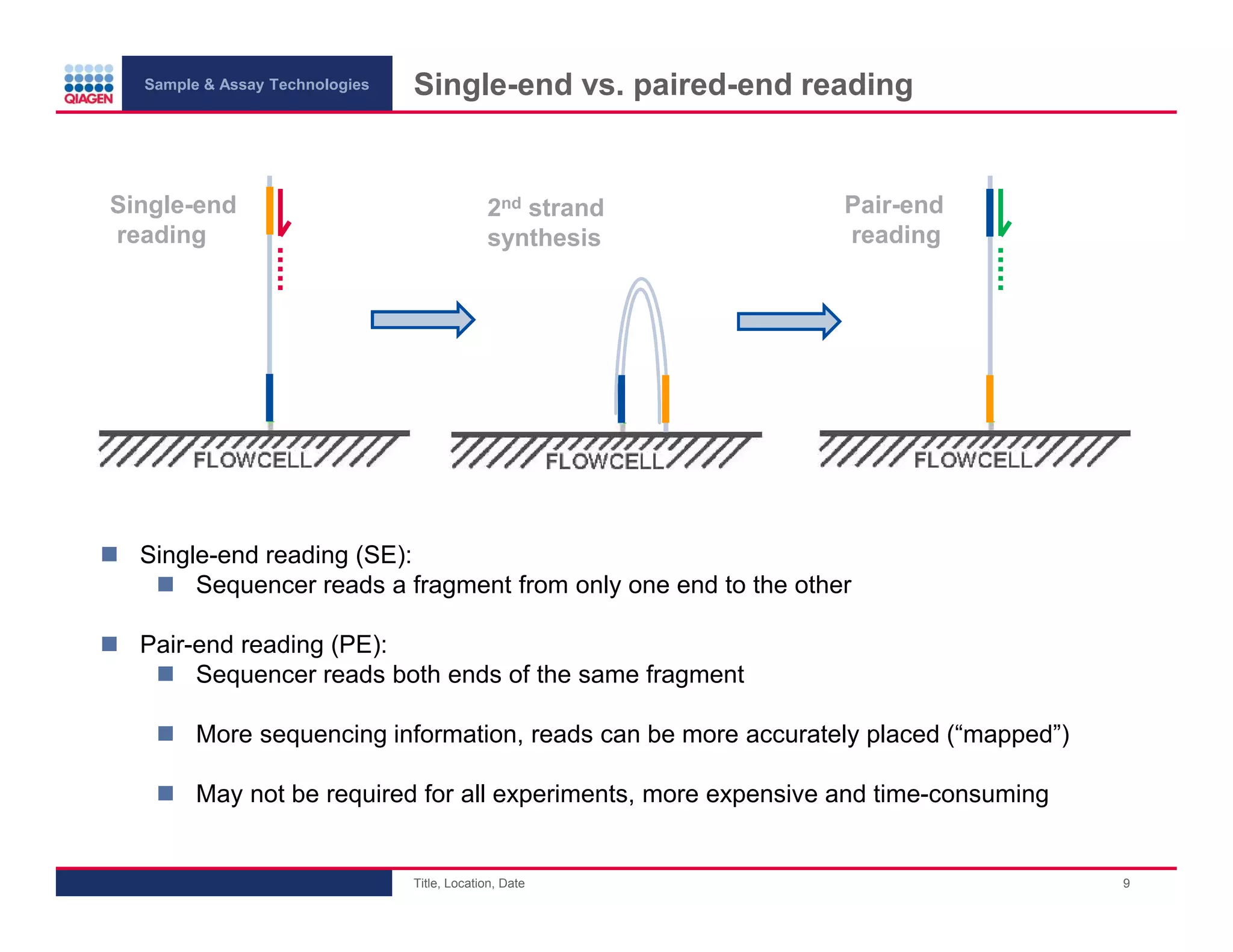
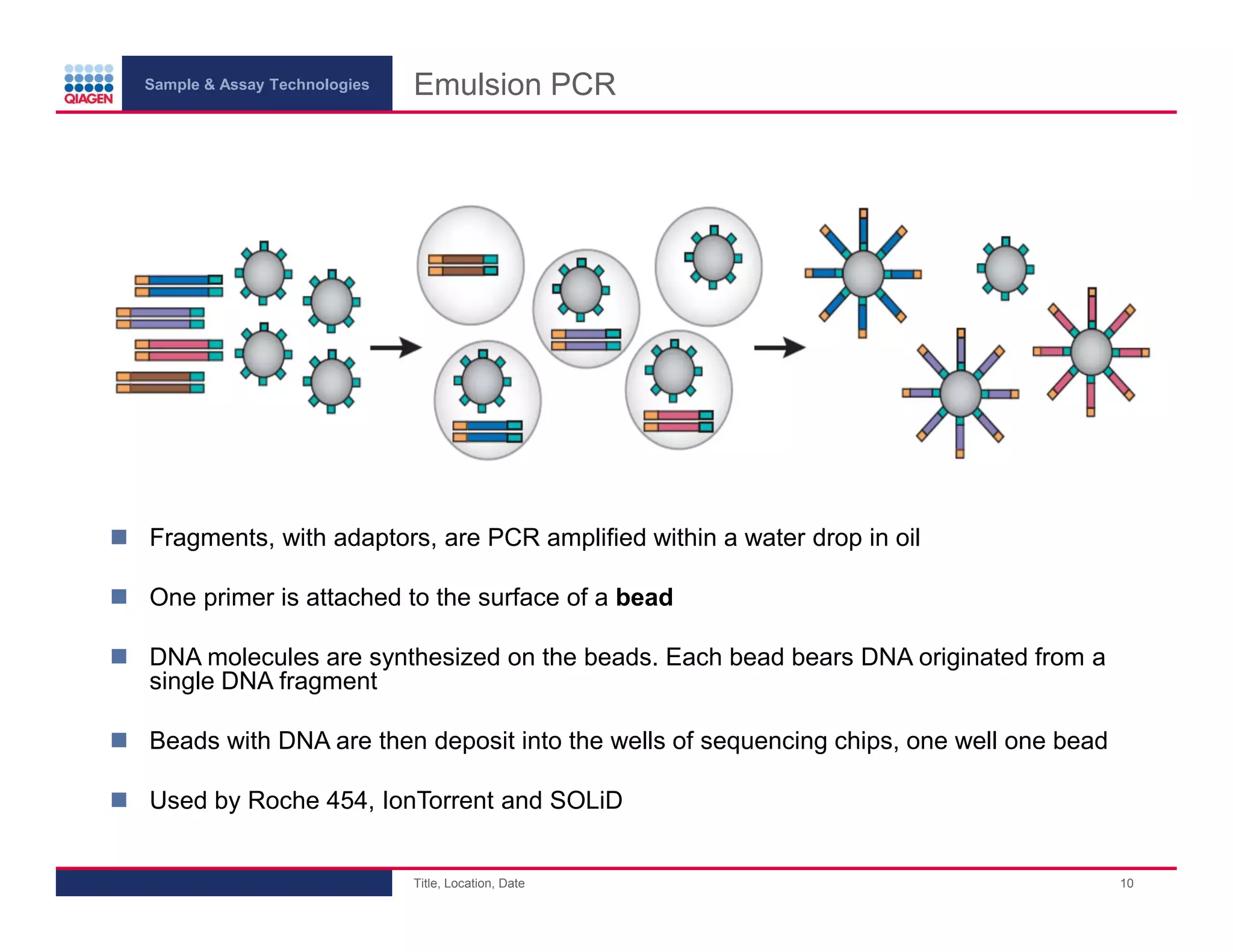
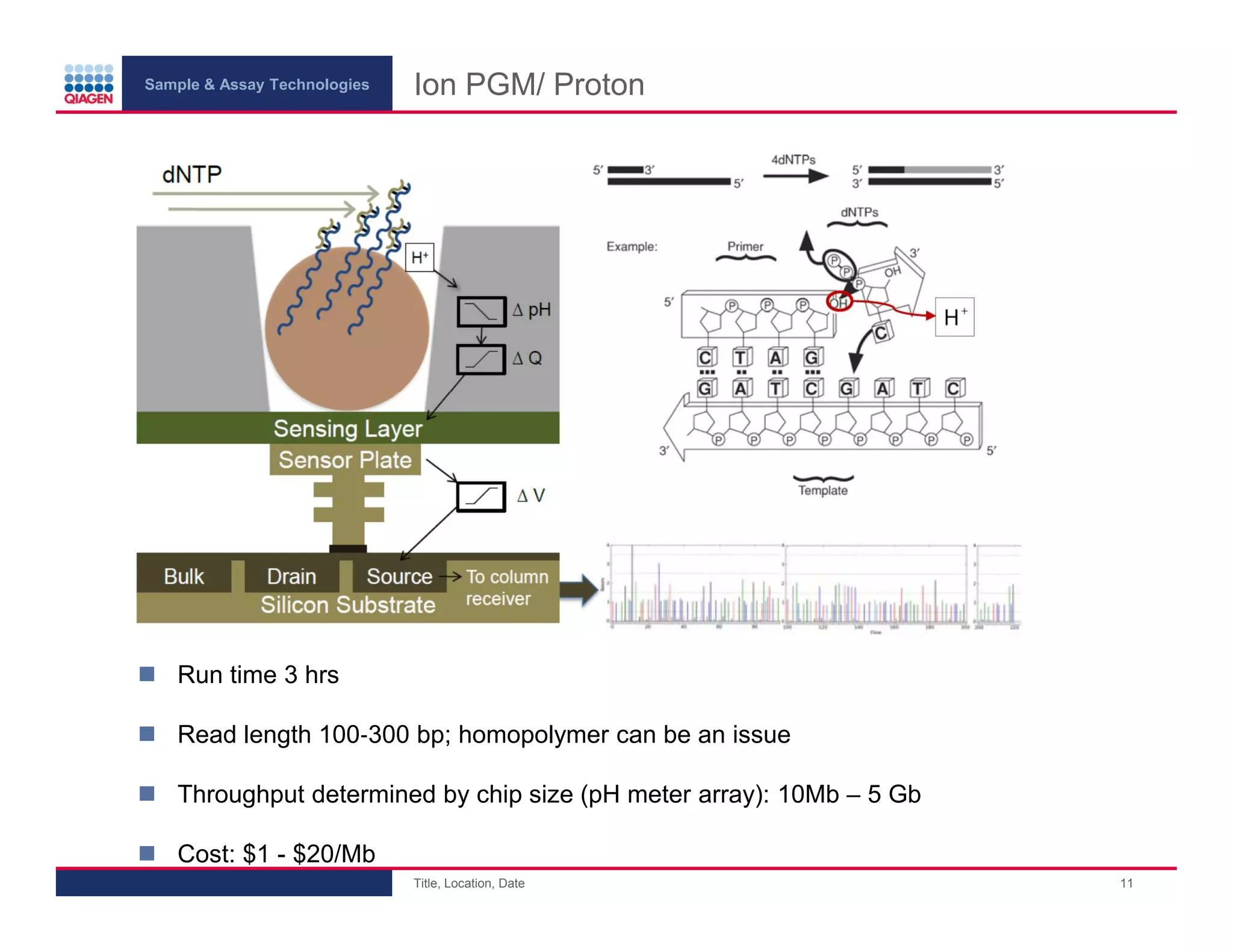
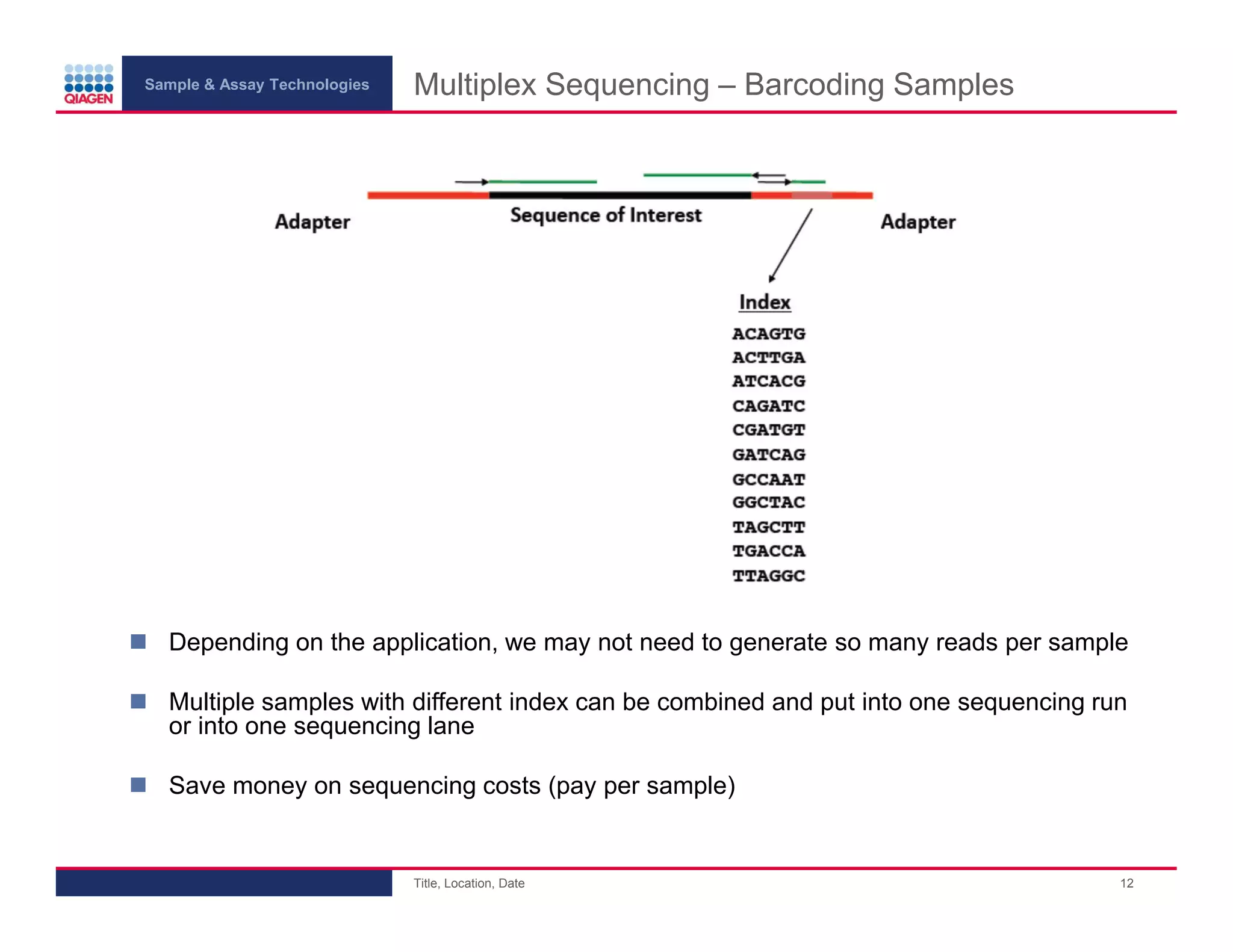
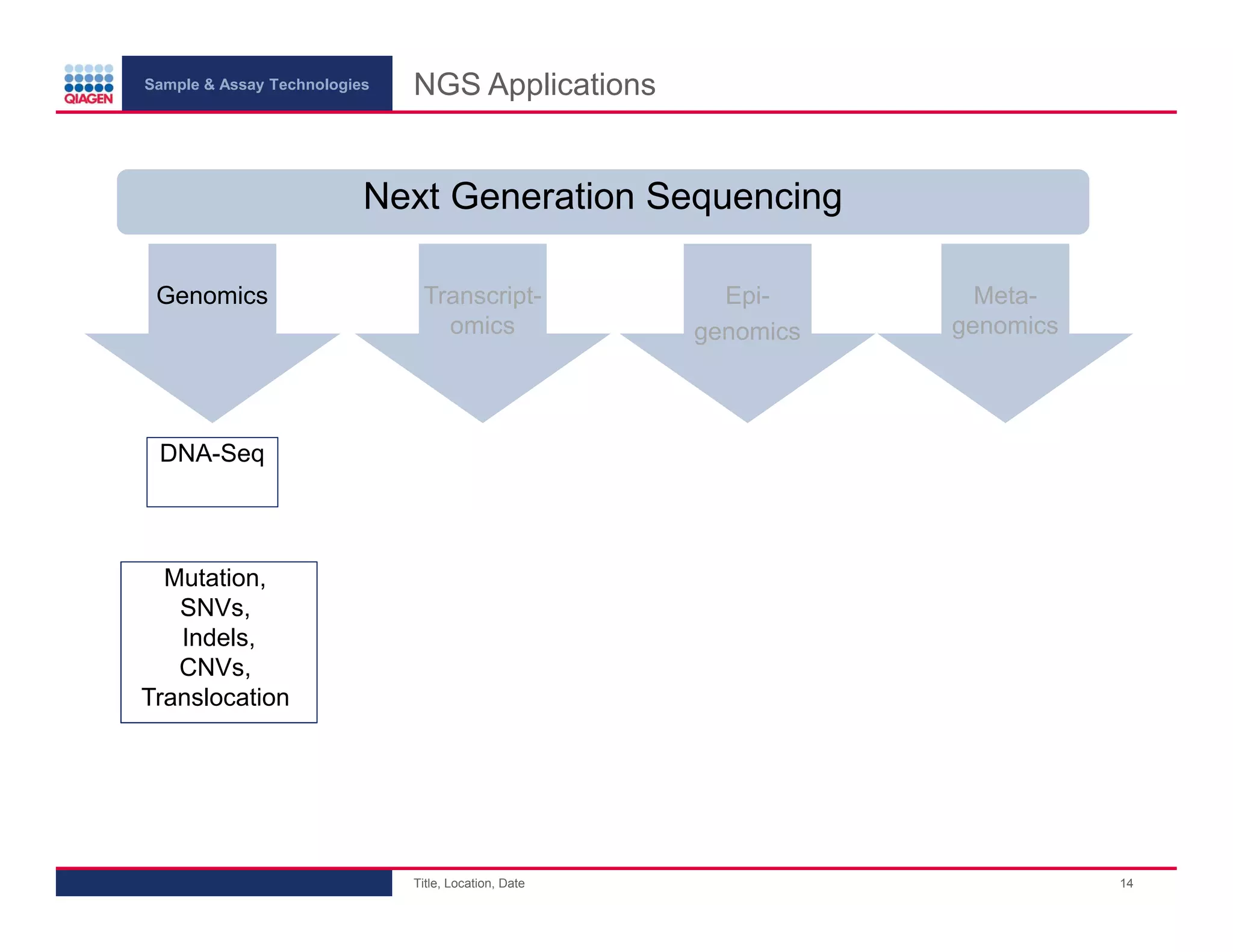
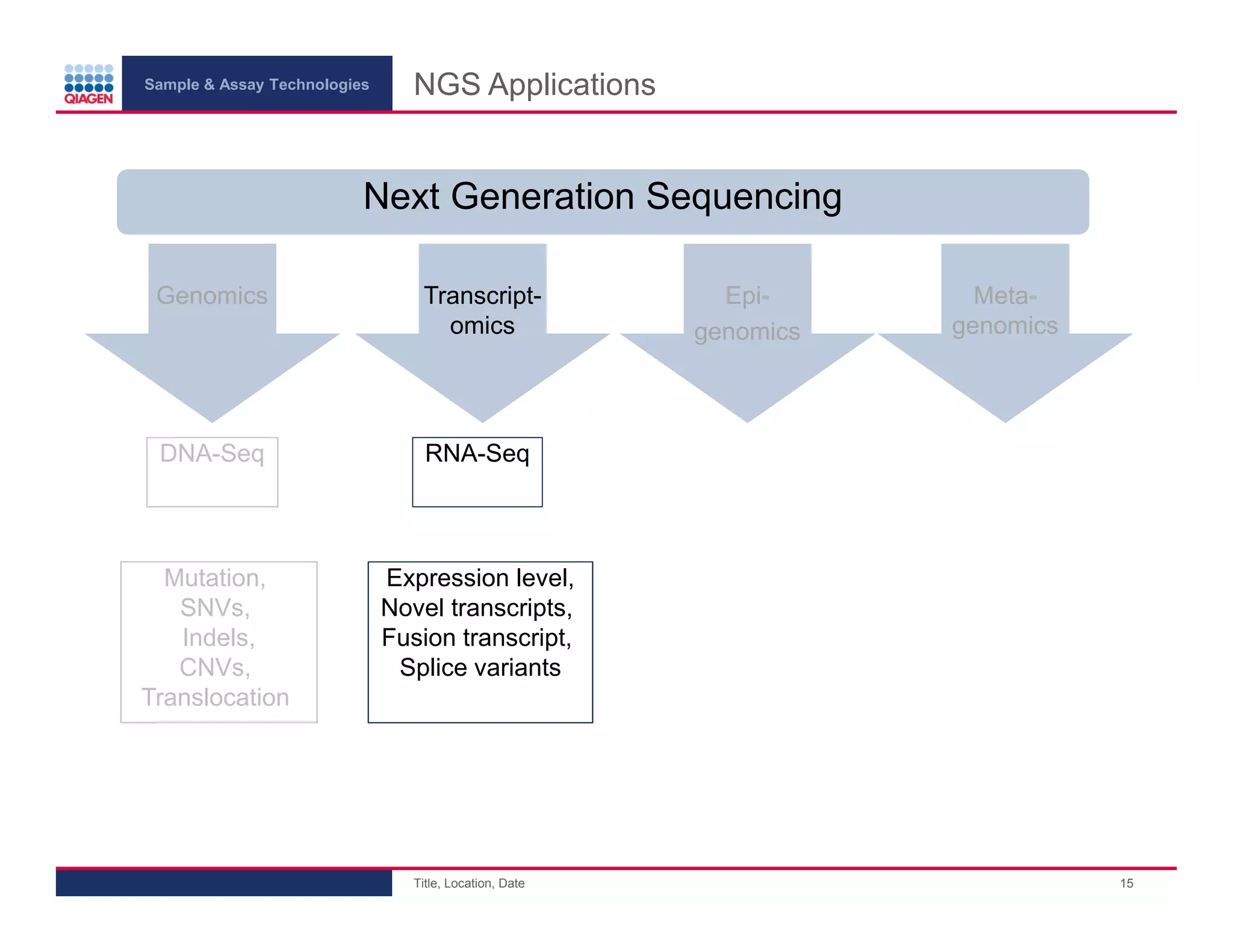
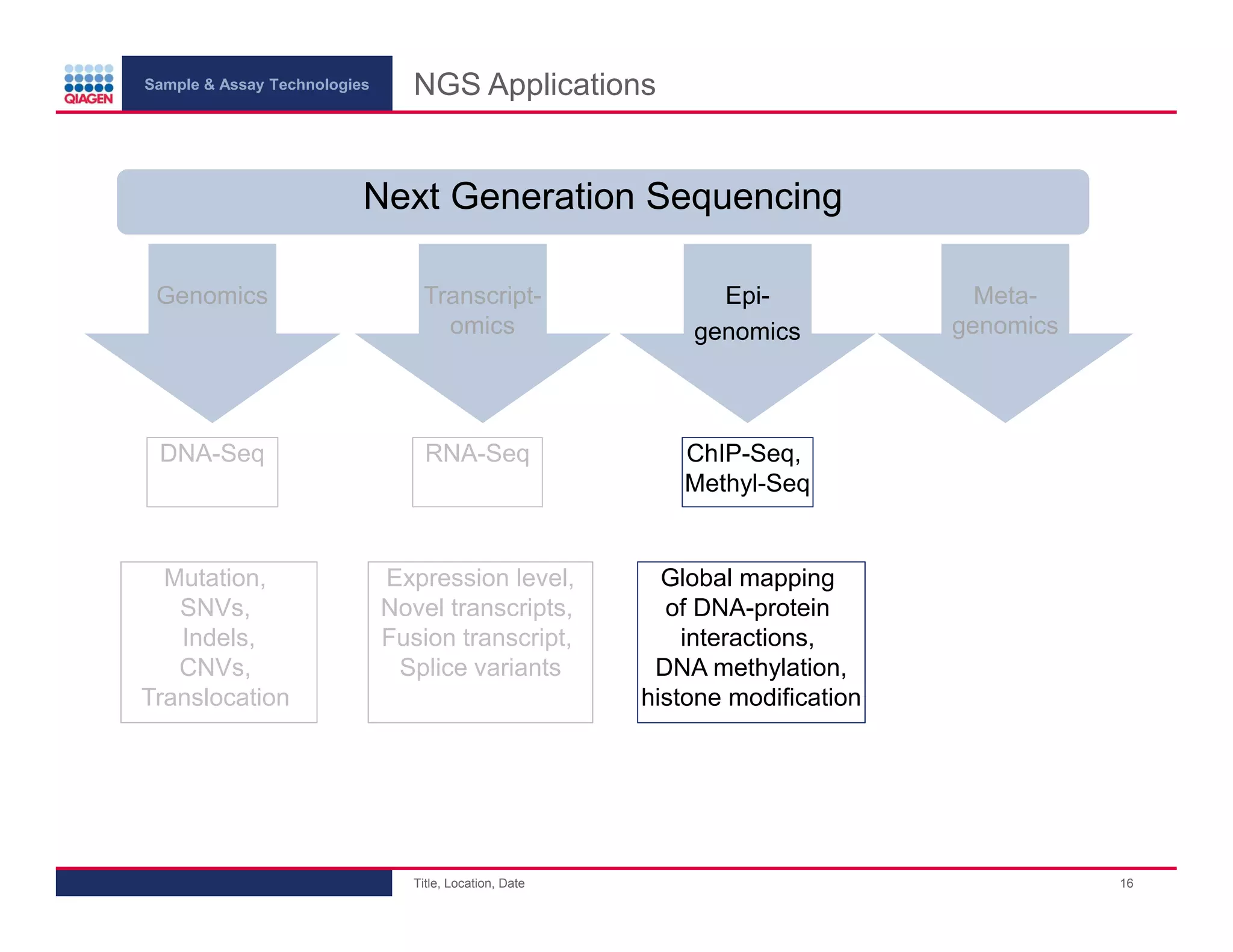
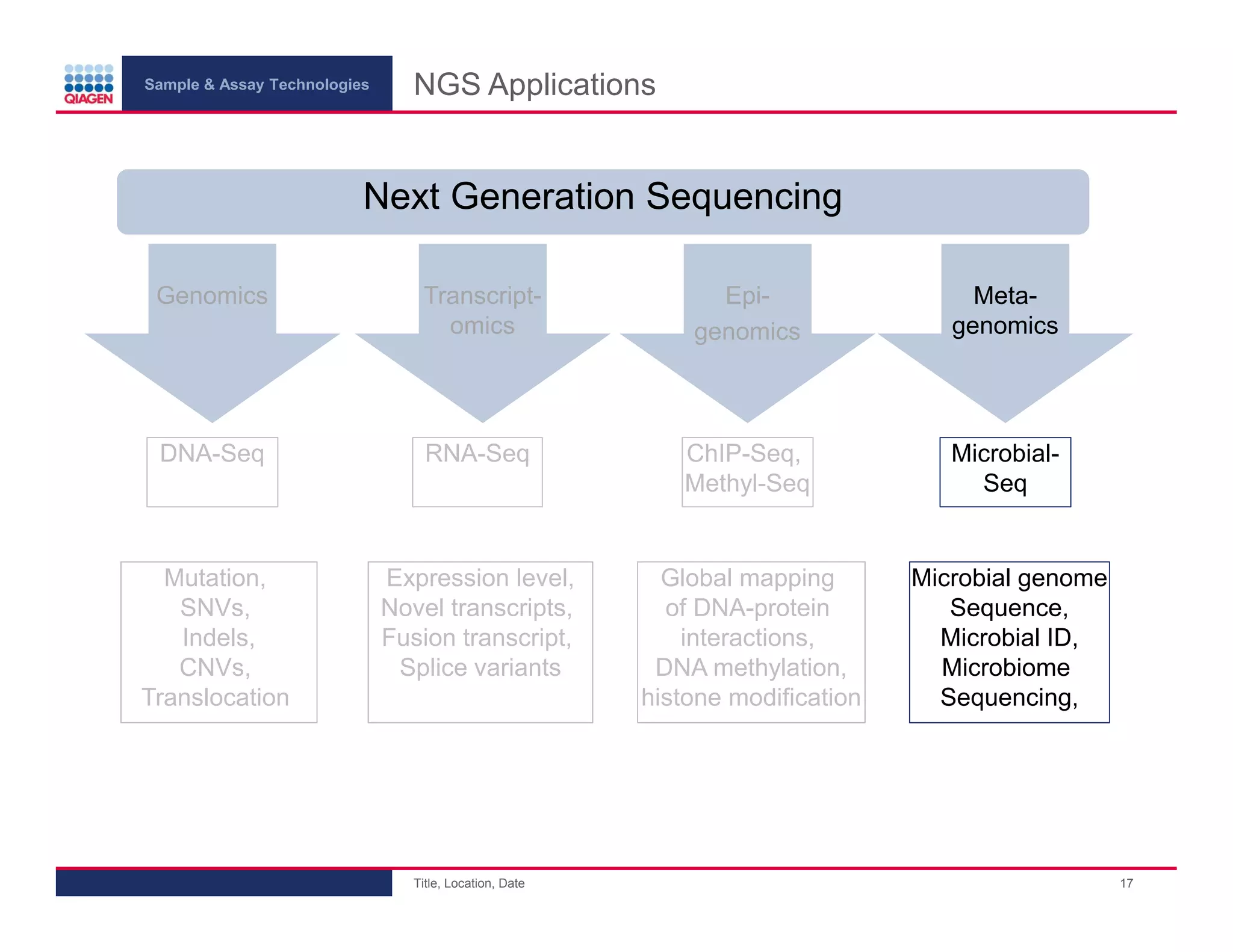
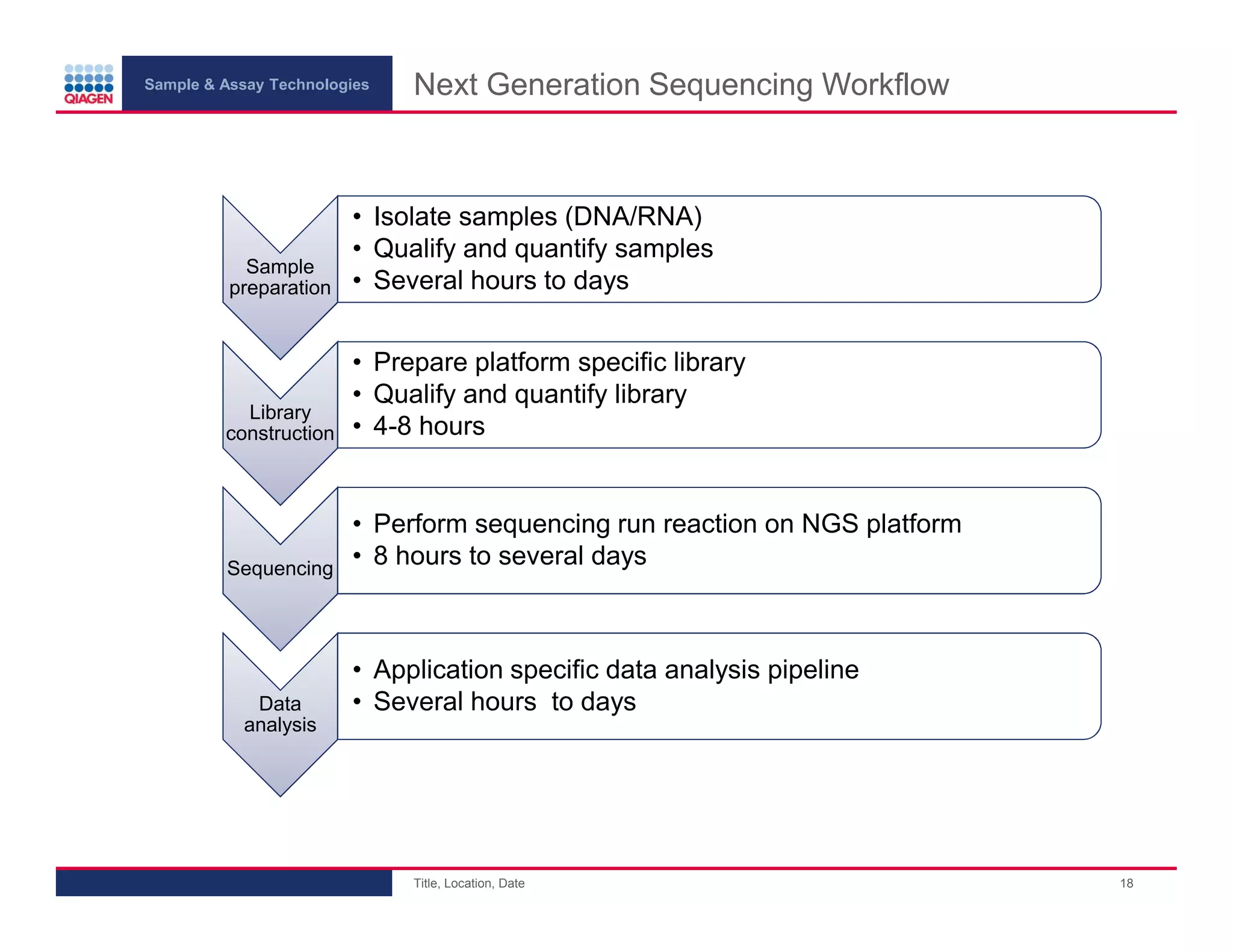
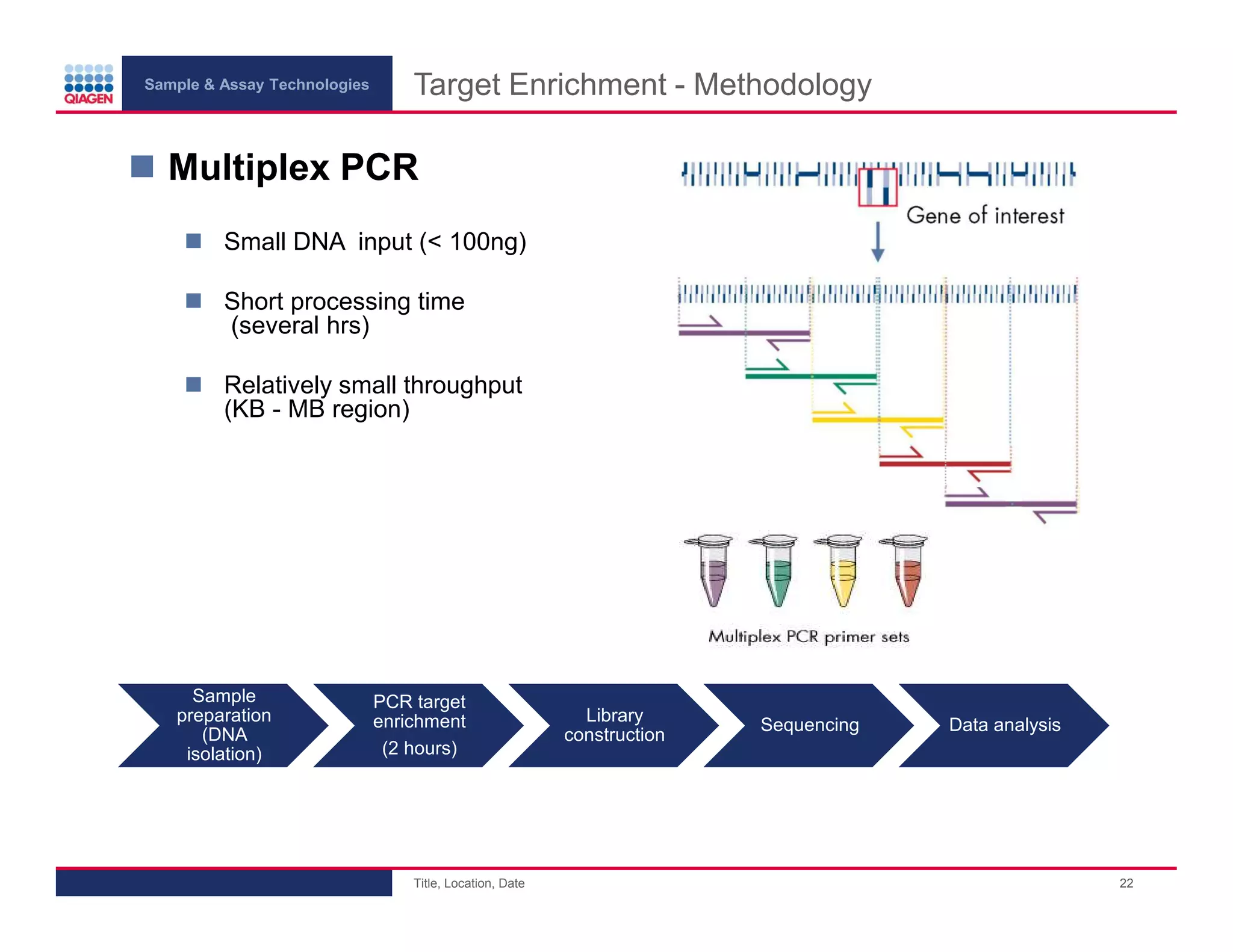
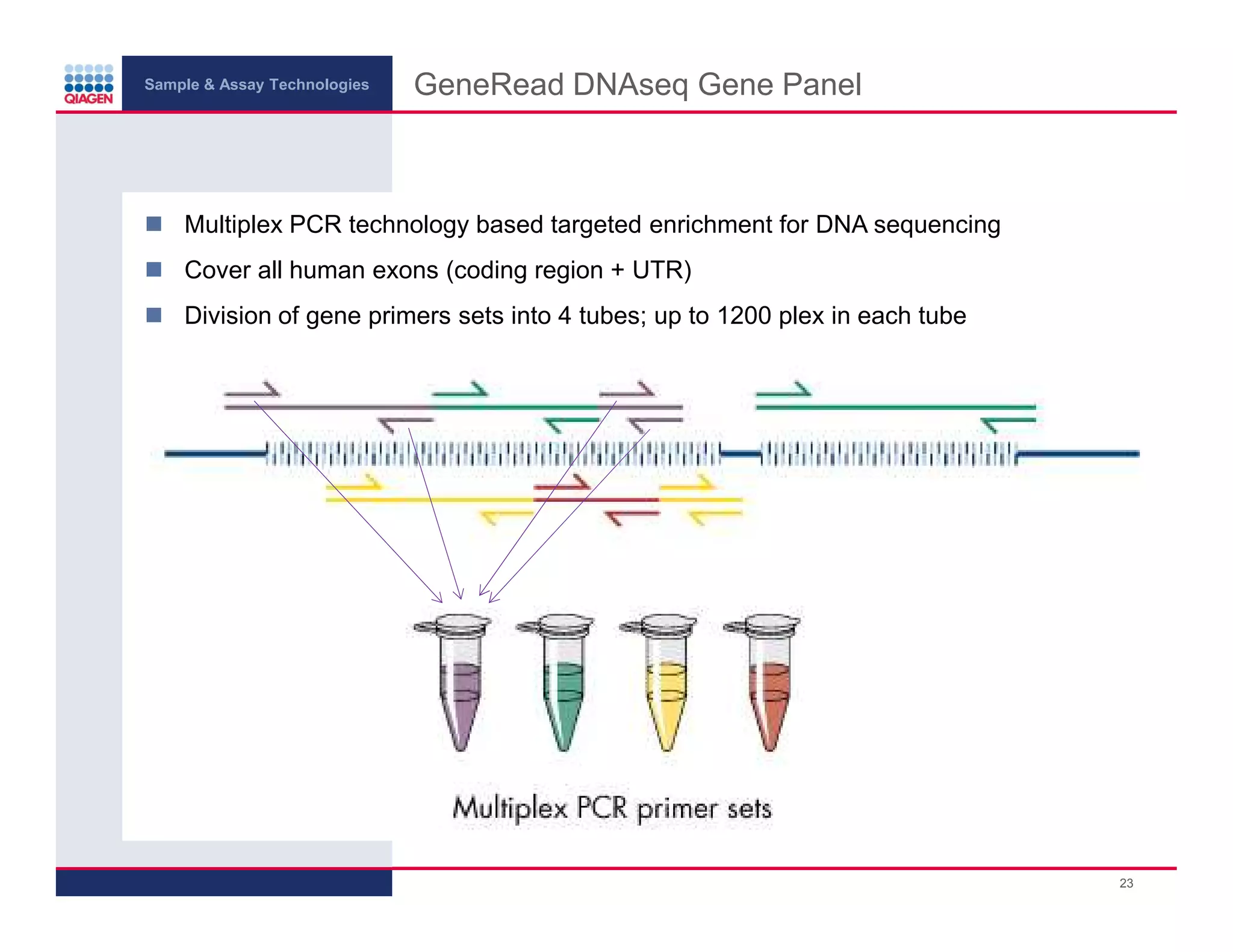
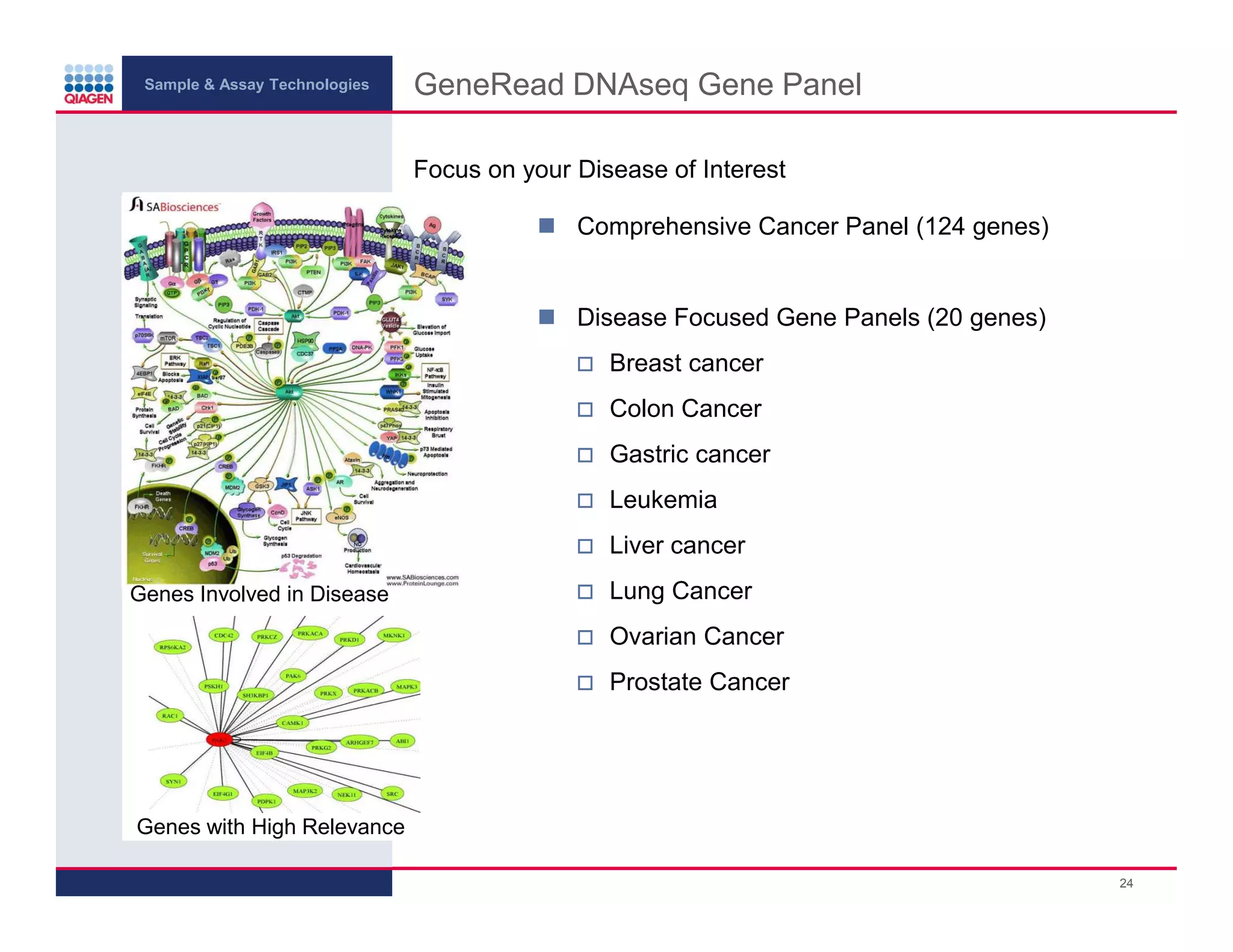
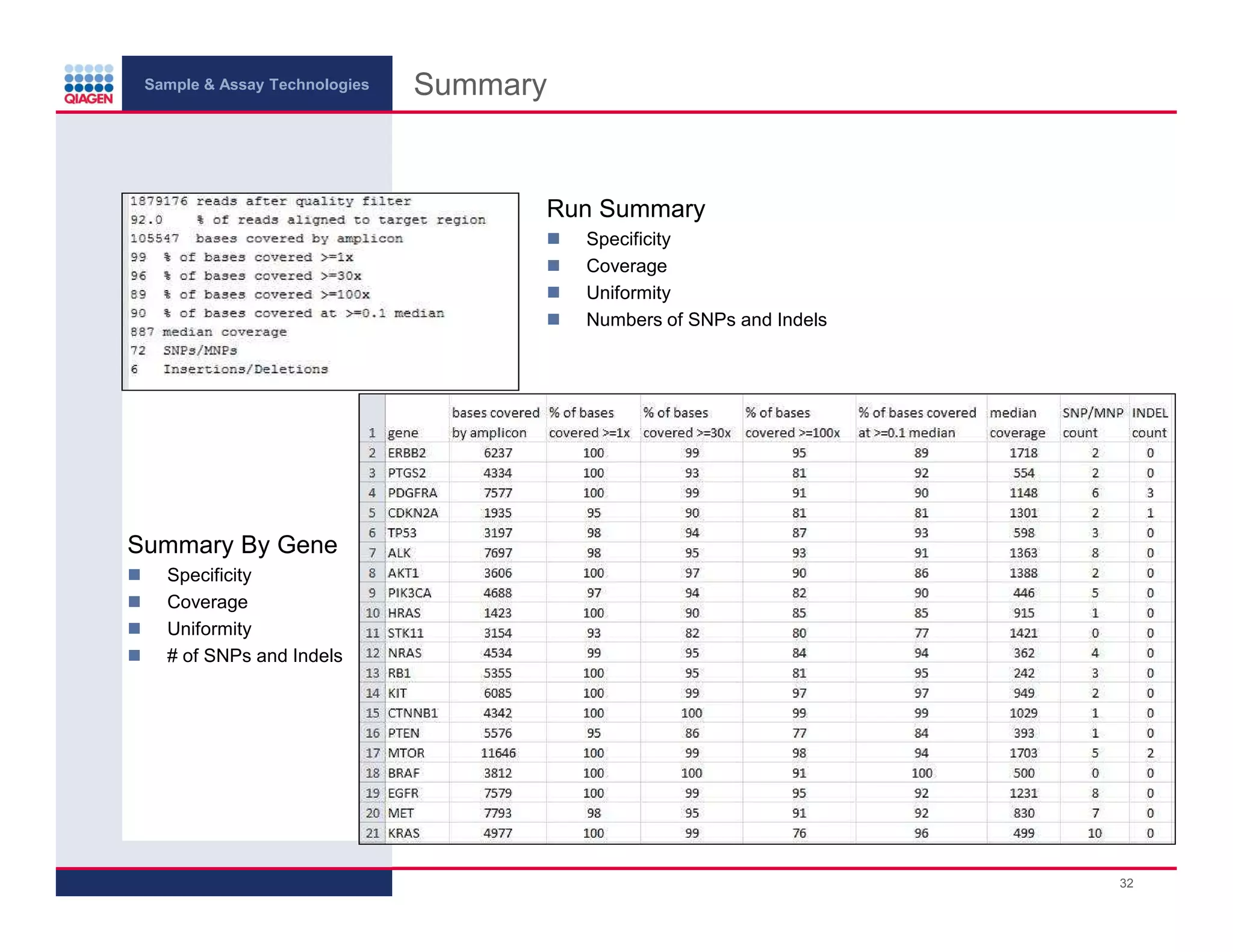
This document provides an overview of next generation sequencing technologies and applications. It summarizes an upcoming webinar series on next generation sequencing and its role in cancer biology. The first webinar will provide an introduction to next generation sequencing technologies and applications and be presented by Quan Peng on April 4, 2013. The following two webinars will focus on next generation sequencing for cancer research and data analysis and be presented on April 11 and 18, 2013 respectively.