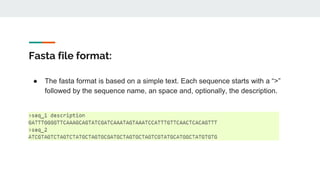
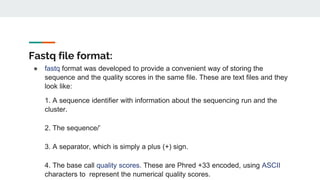
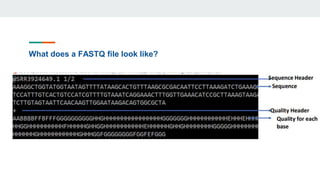
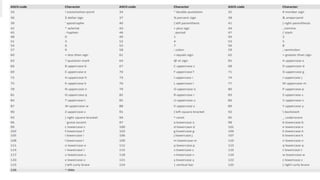
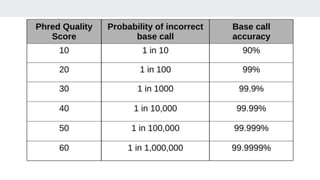
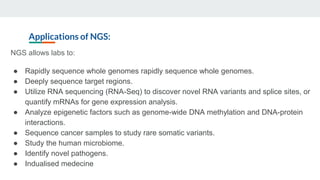
This document provides an overview of next generation sequencing (NGS), including common file formats like FASTA, FASTQ, and SRA. It discusses the applications of NGS such as genome sequencing, RNA sequencing, and microbiome analysis. Finally, it outlines some drawbacks of NGS, such as the unknown clinical significance of some identified variants and the large data storage requirements.