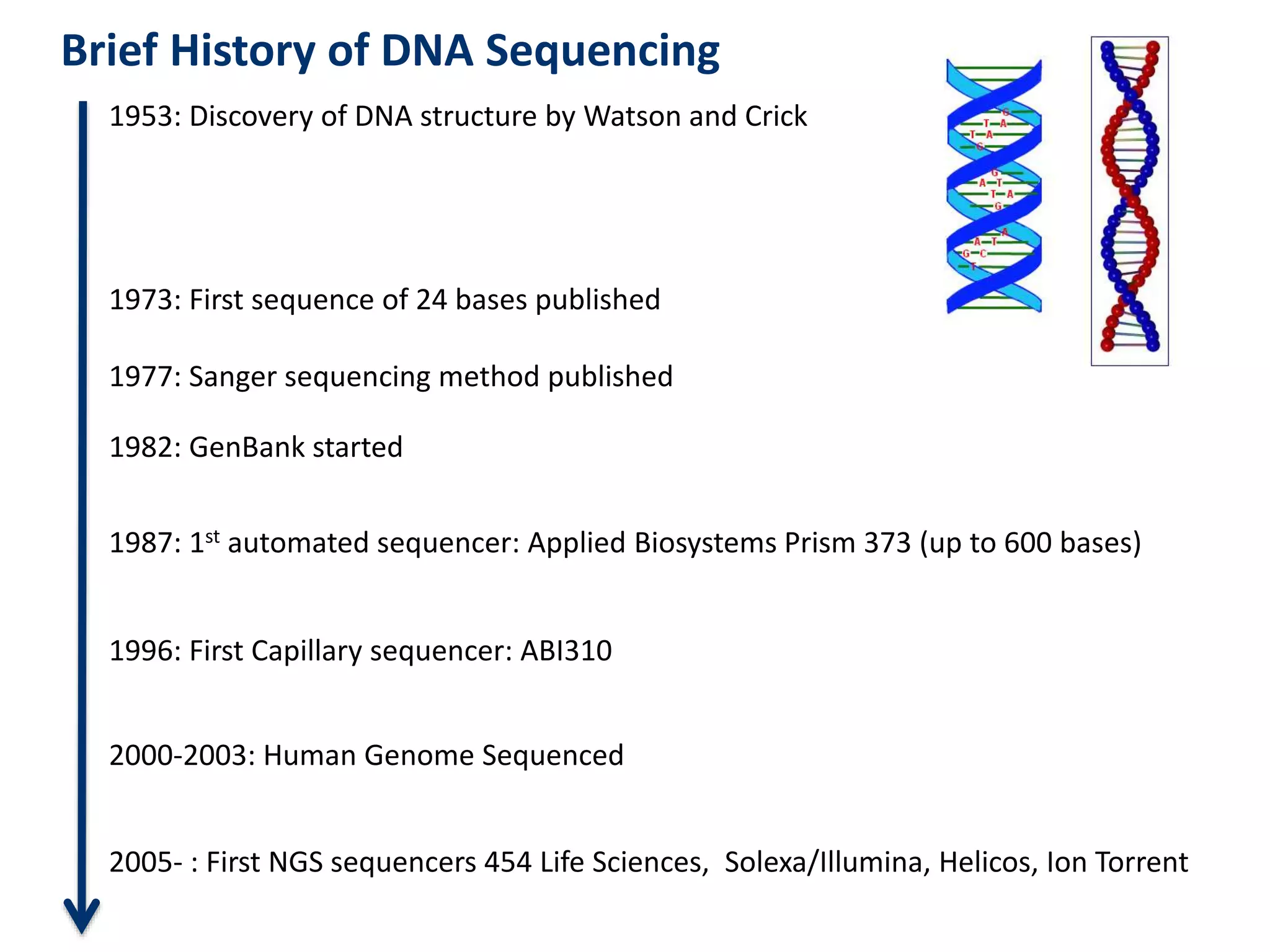
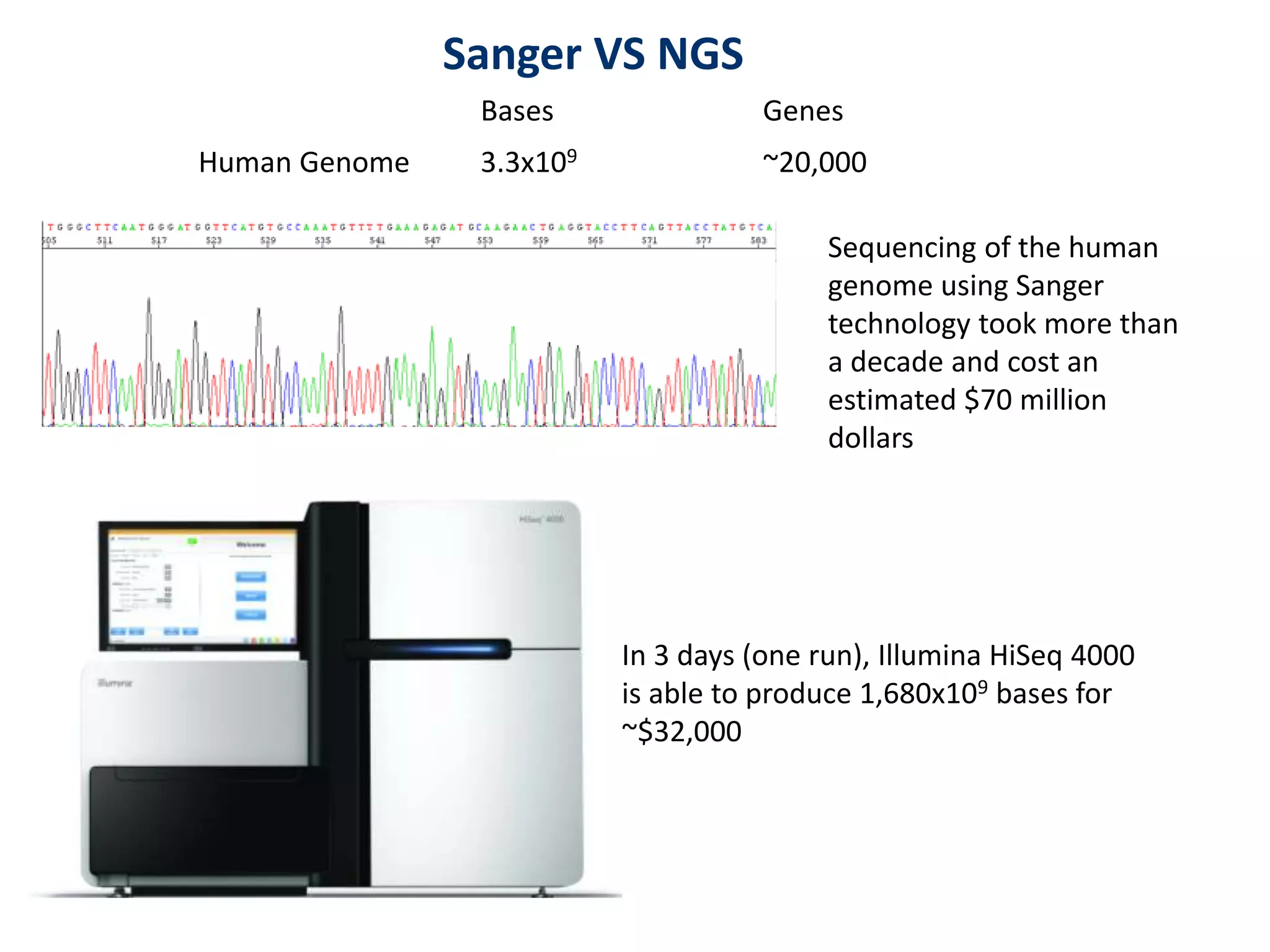
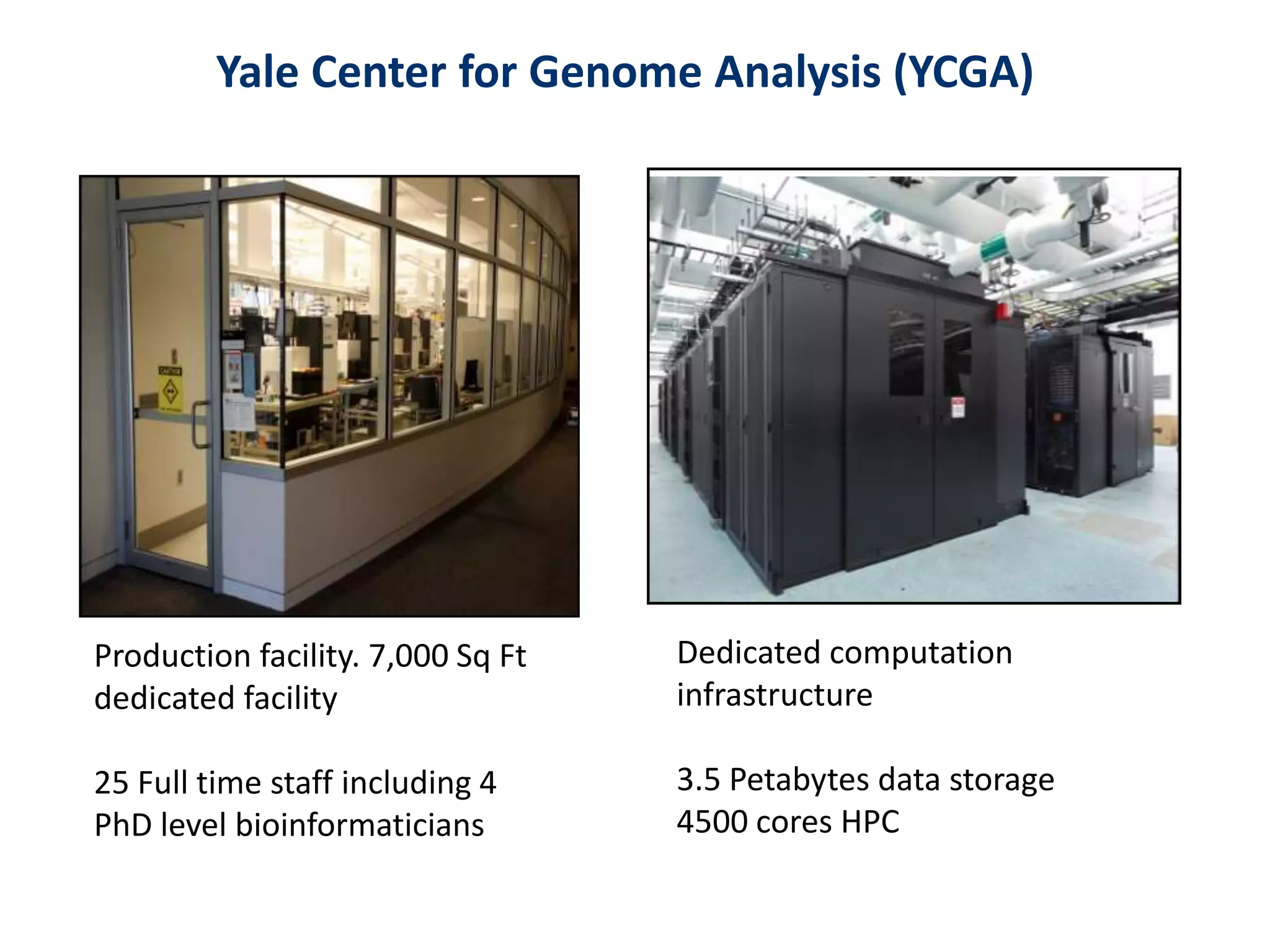
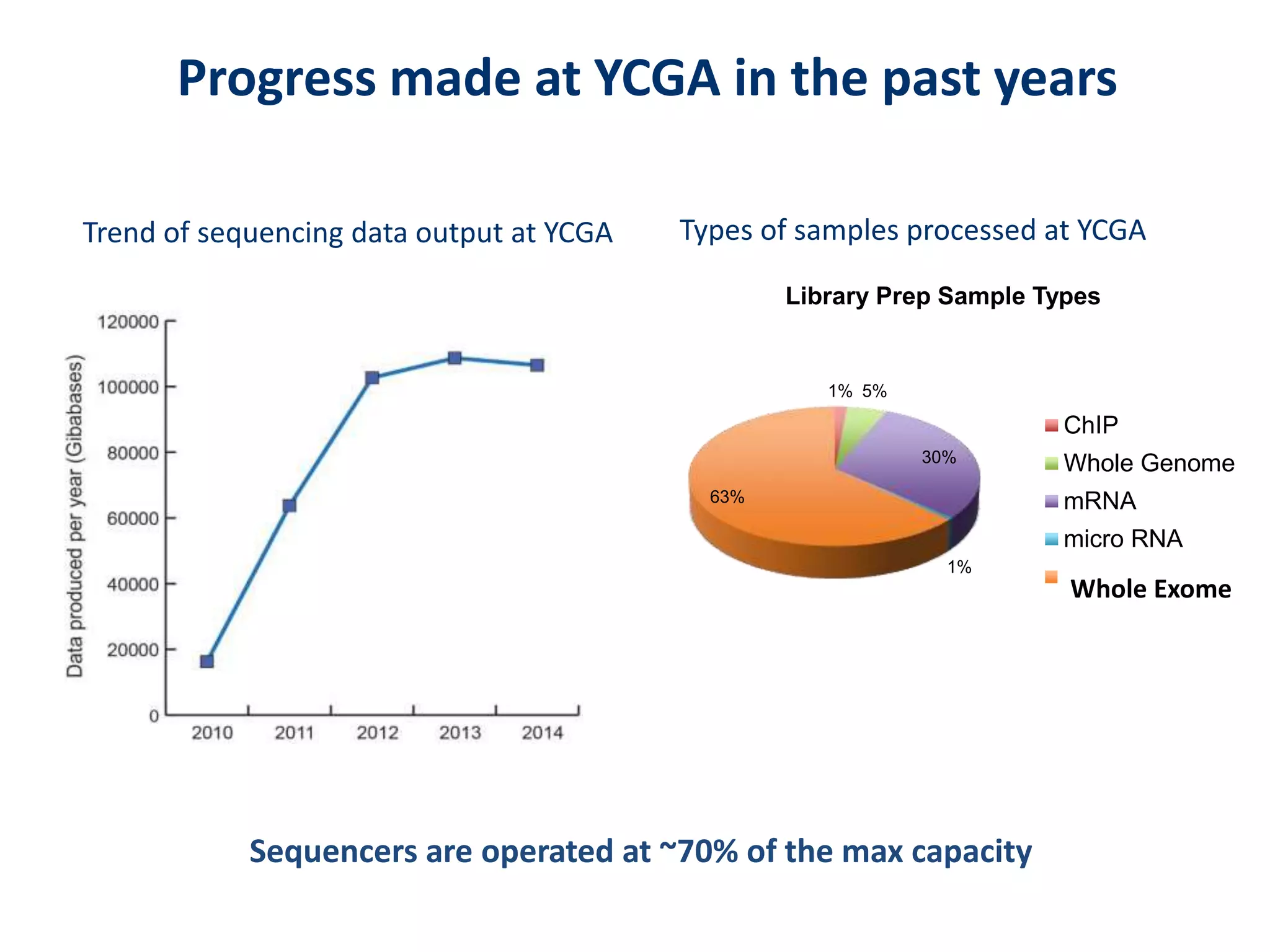
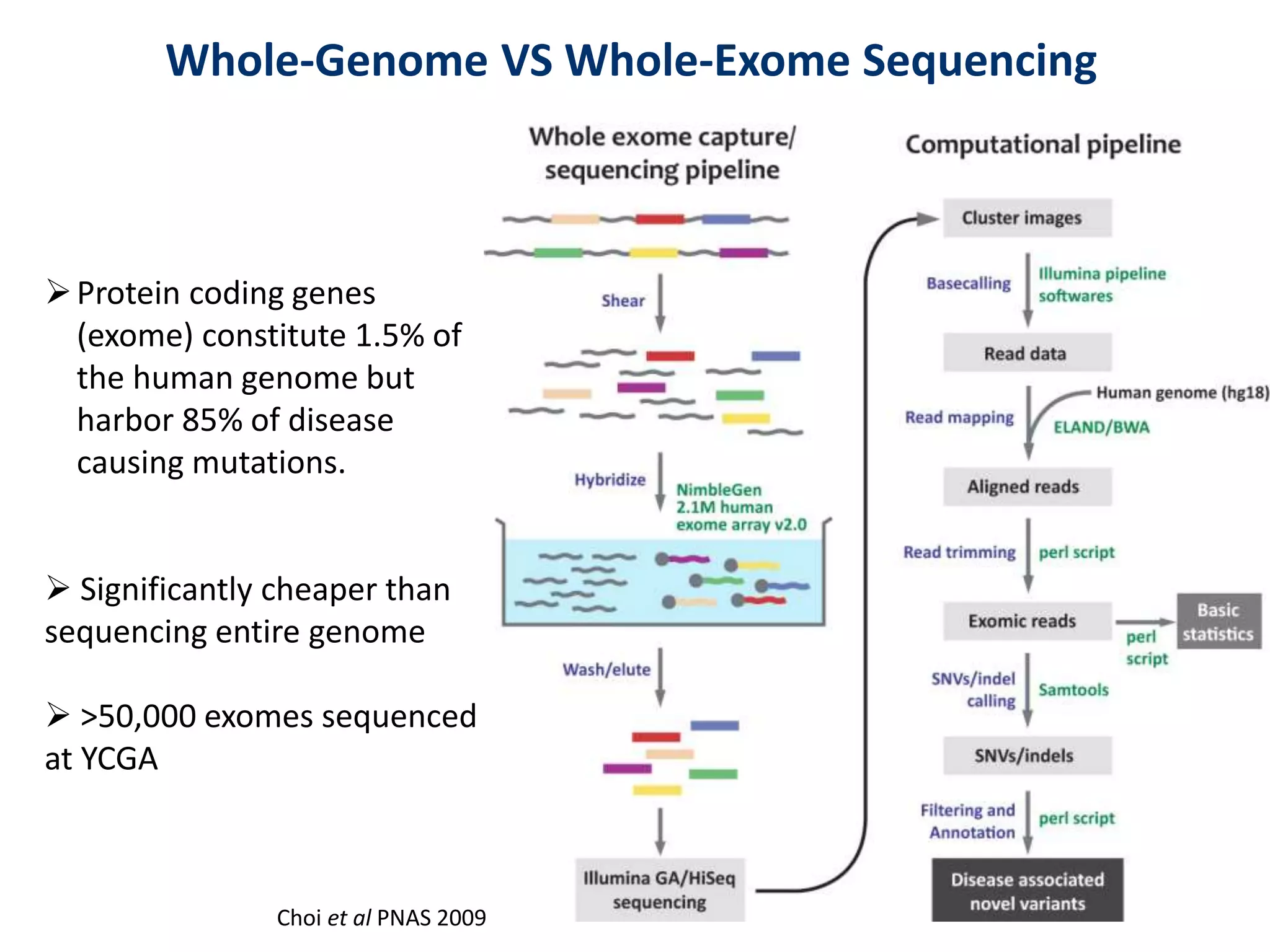
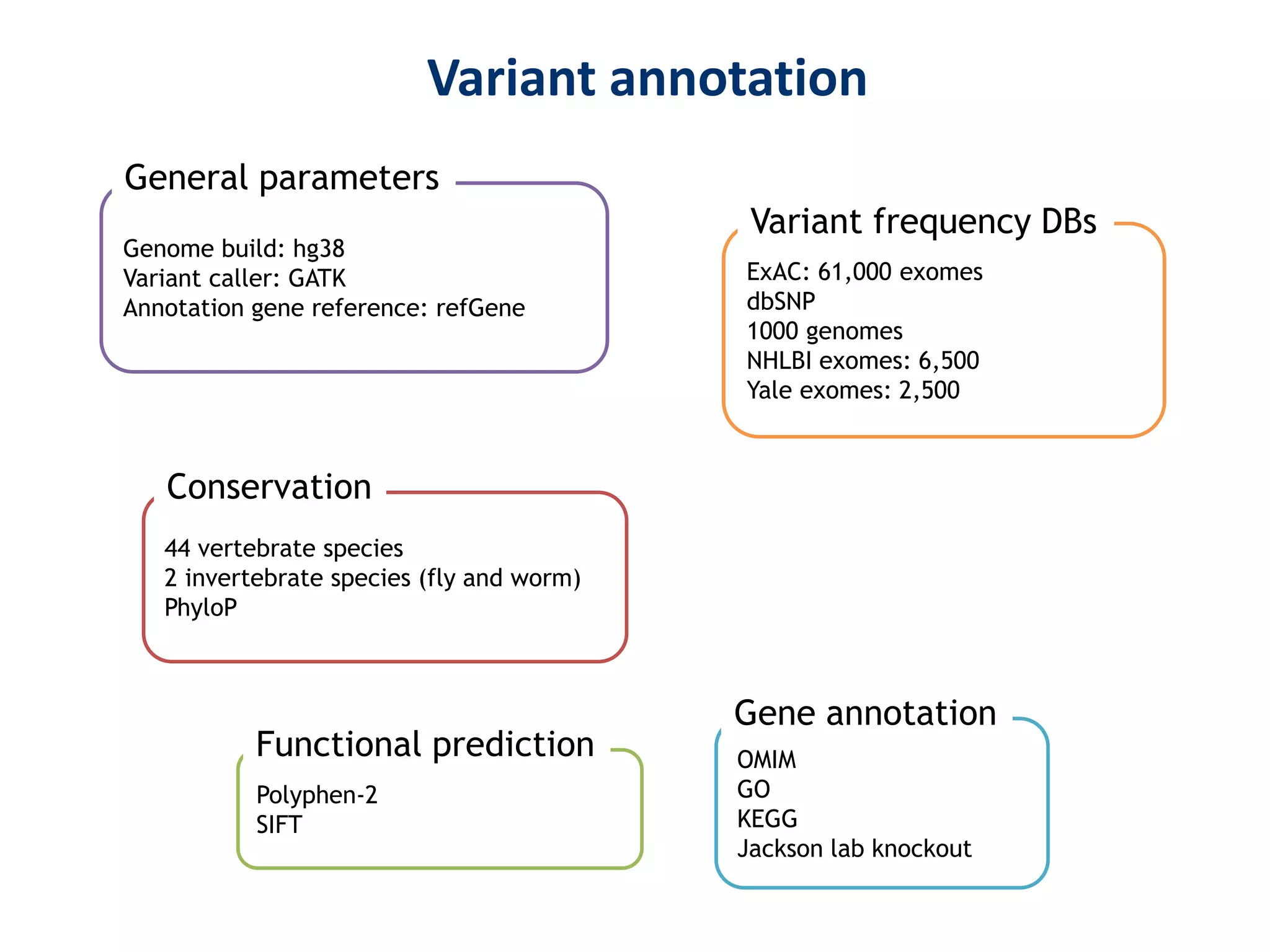
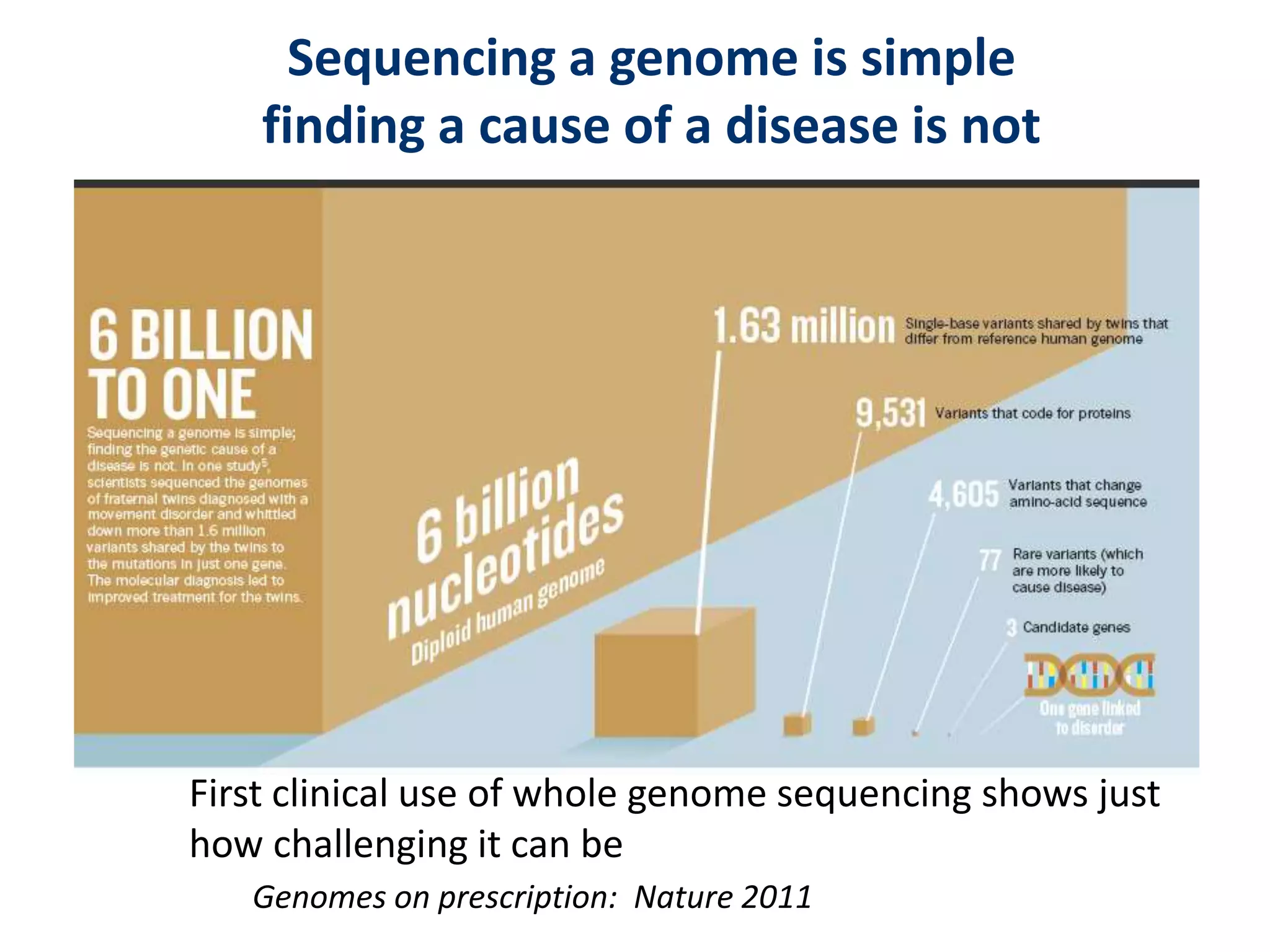
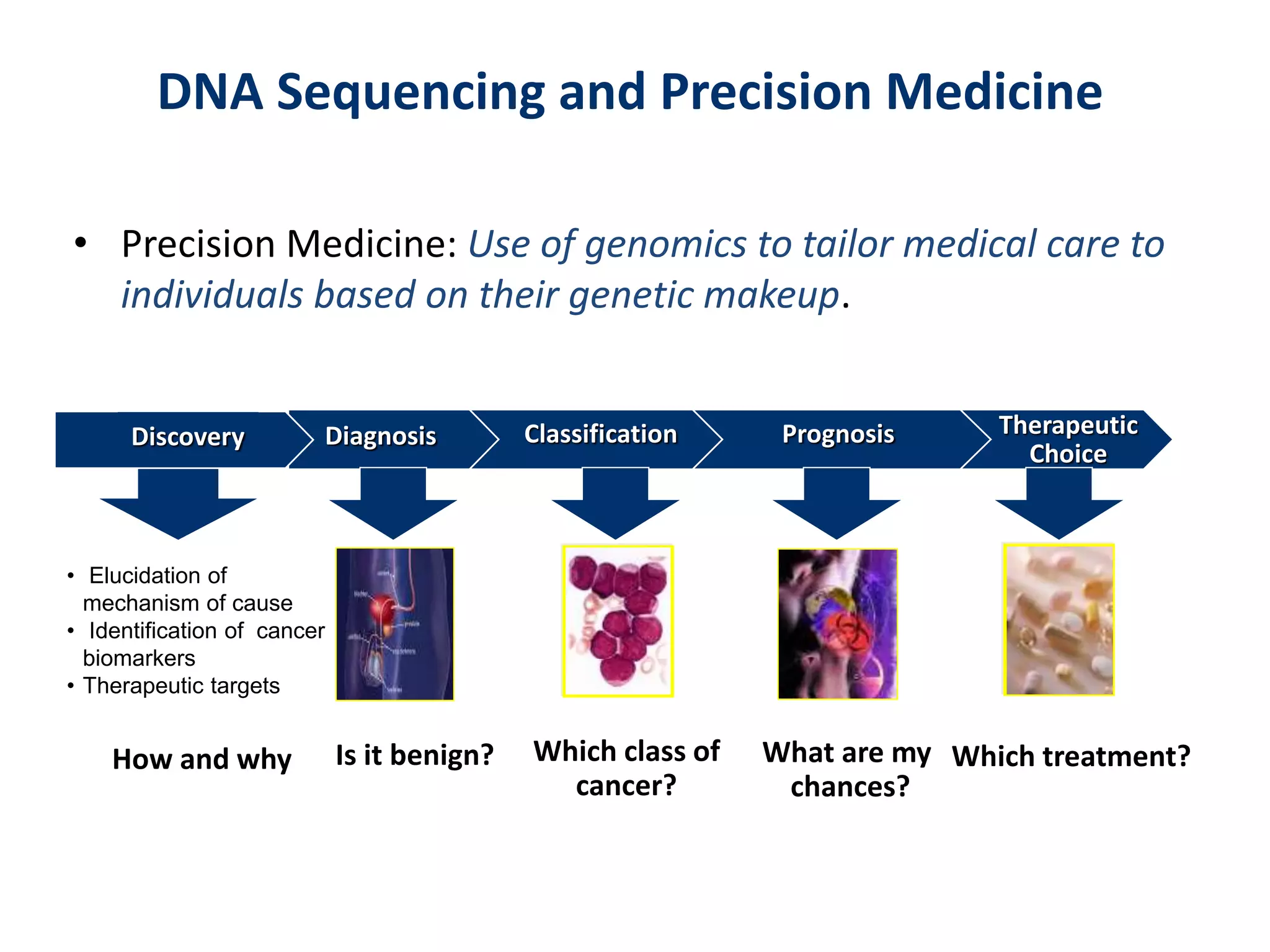
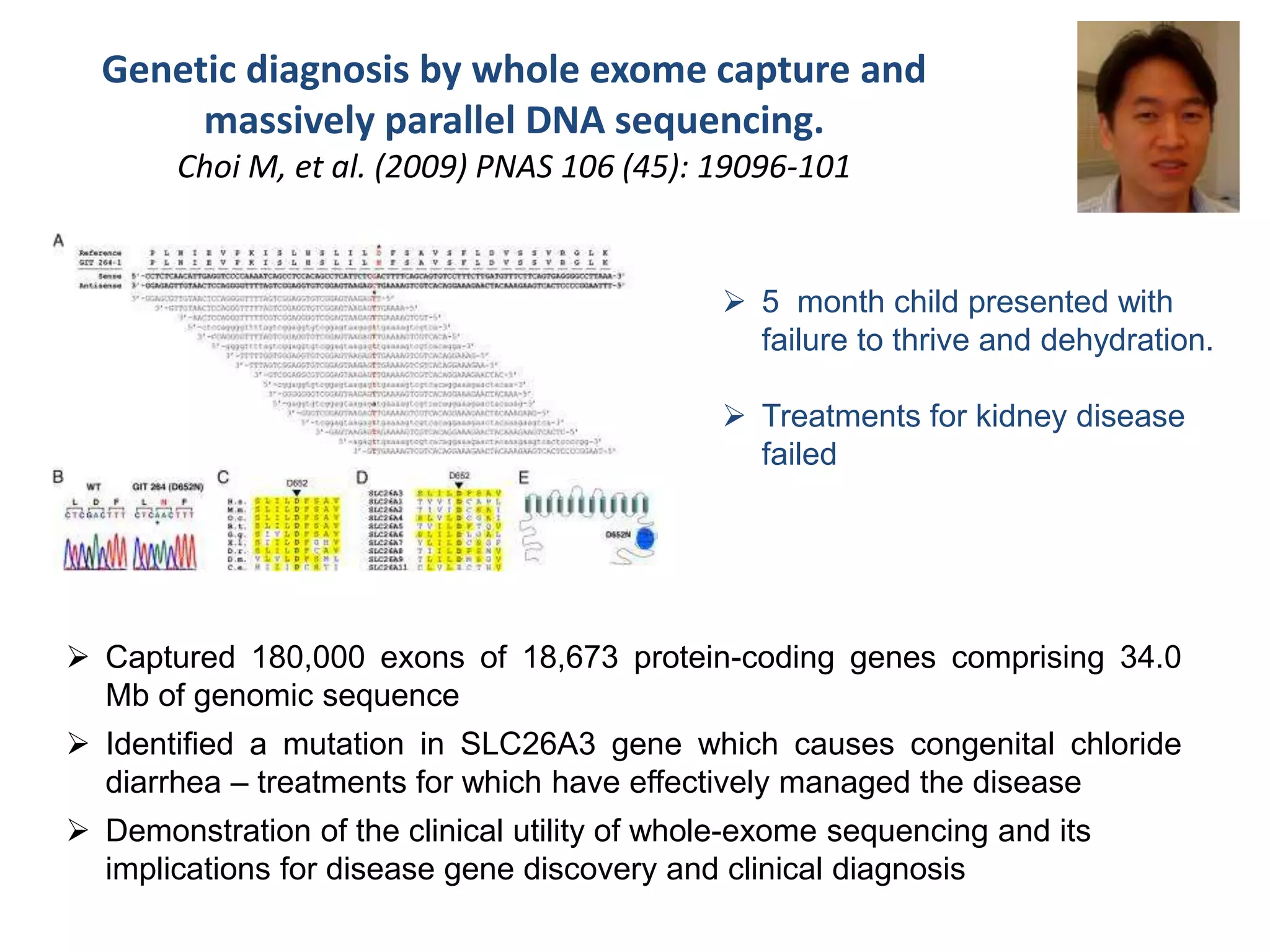
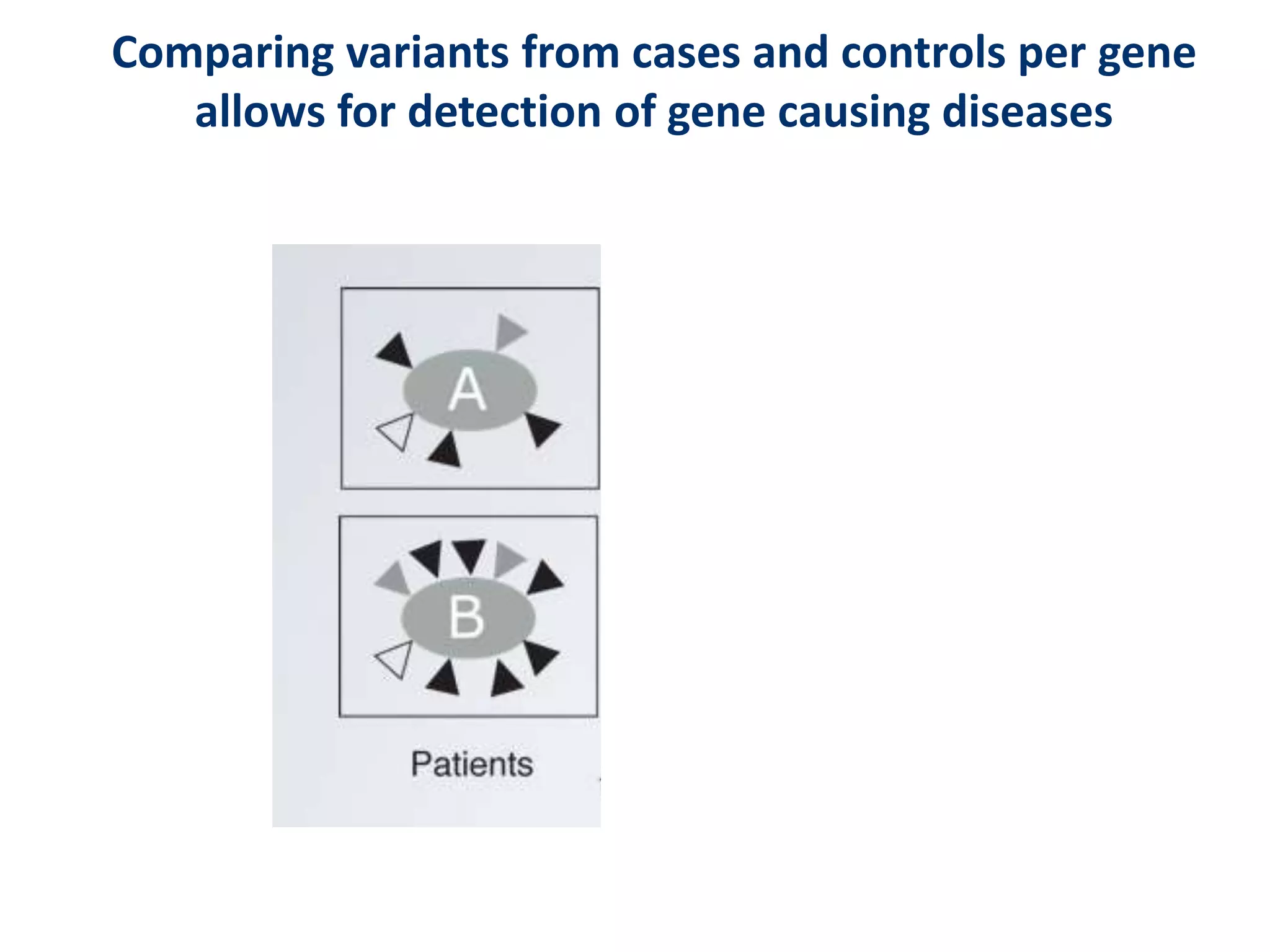
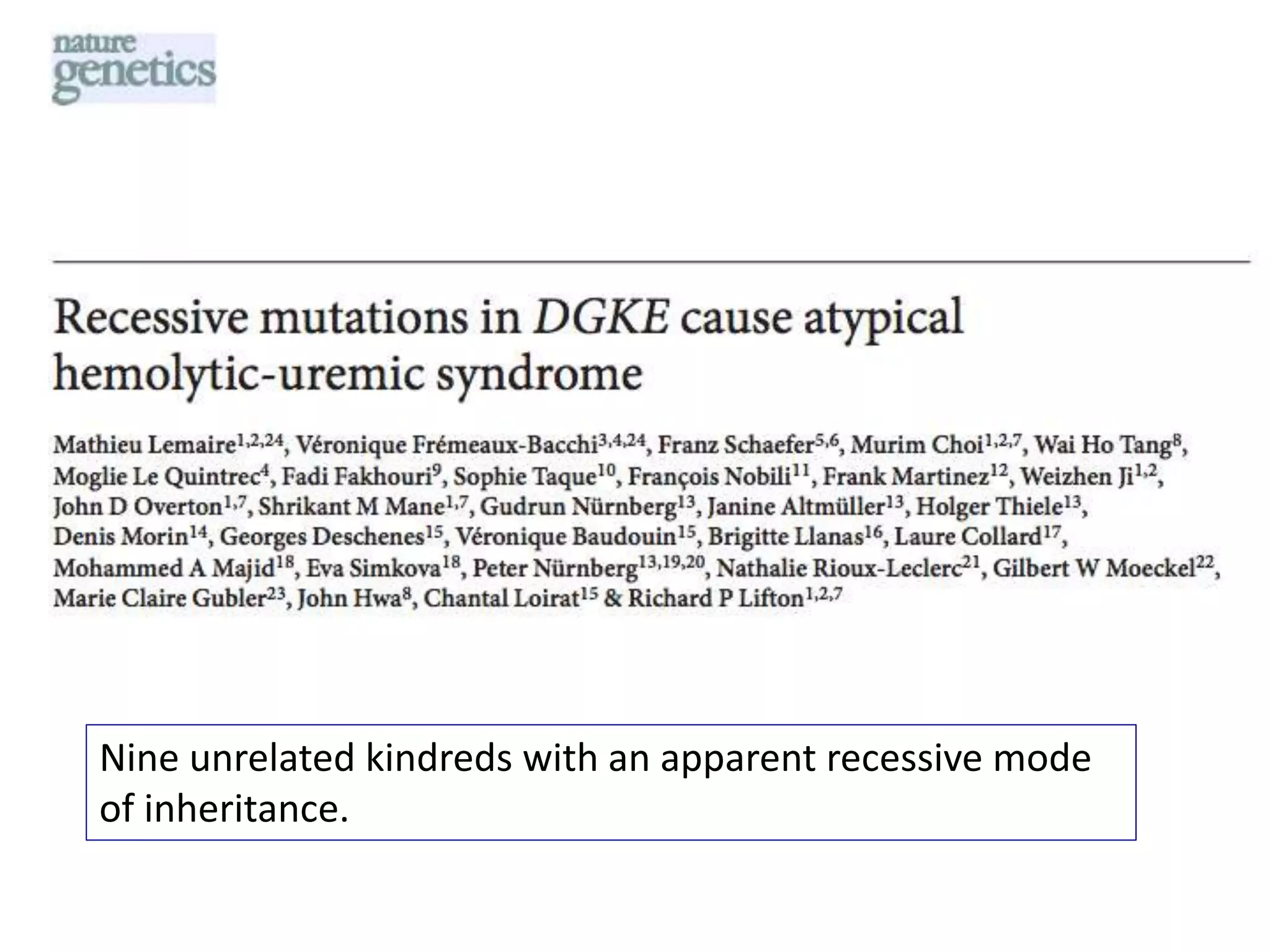
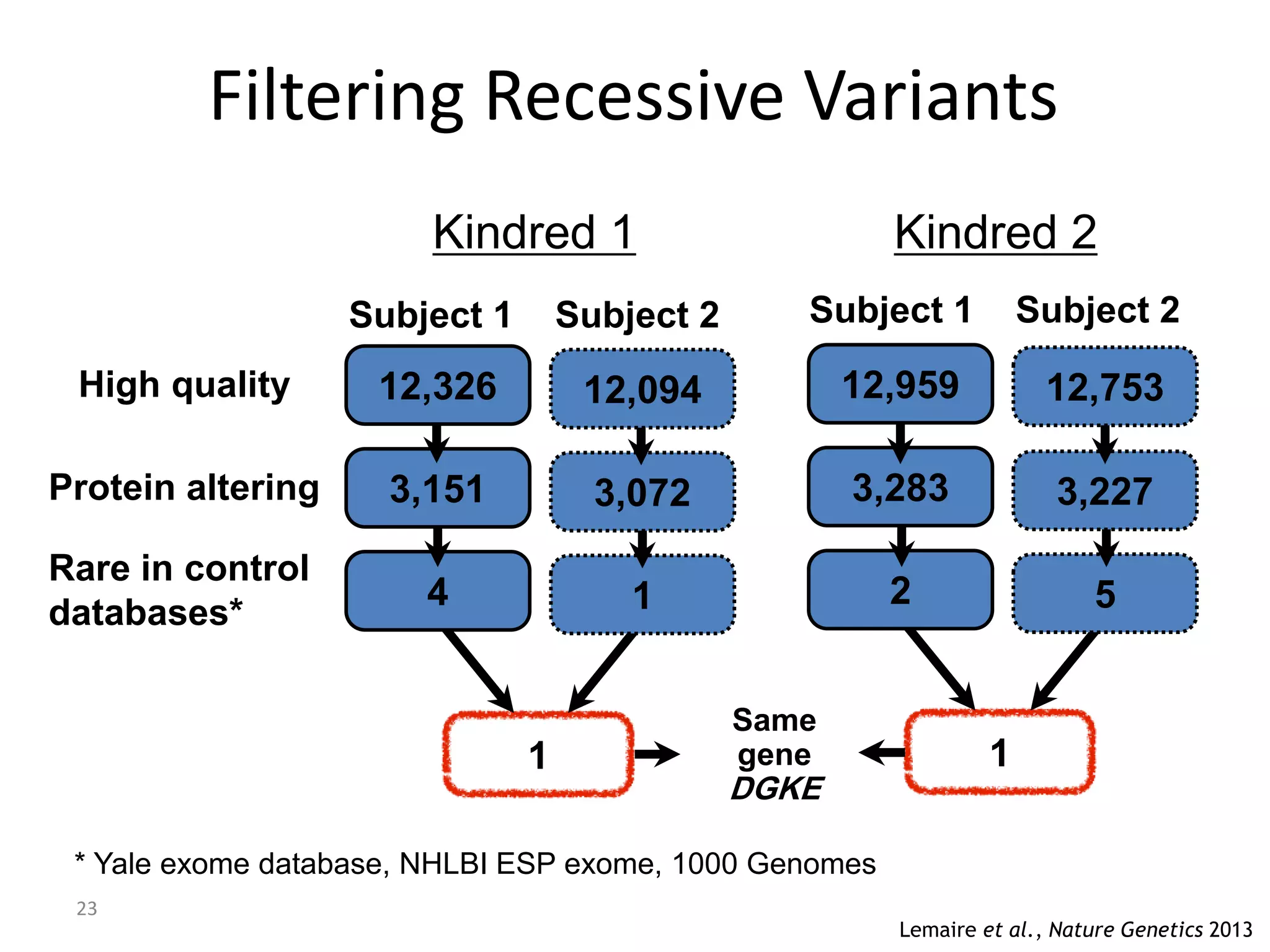
The document outlines the history and advancements in DNA sequencing, focusing on next-generation sequencing (NGS) technology and its applications in medical research, particularly at the Yale Center for Genome Analysis. It discusses the evolution of sequencing methods, the efficiency of whole-exome versus whole-genome sequencing, and highlights significant case studies showcasing the clinical utility of these techniques. Additionally, it touches on the implications of precision medicine through genomics, providing insights into genetic diagnoses and therapeutic targets, alongside a list of notable publications stemming from NGS research.