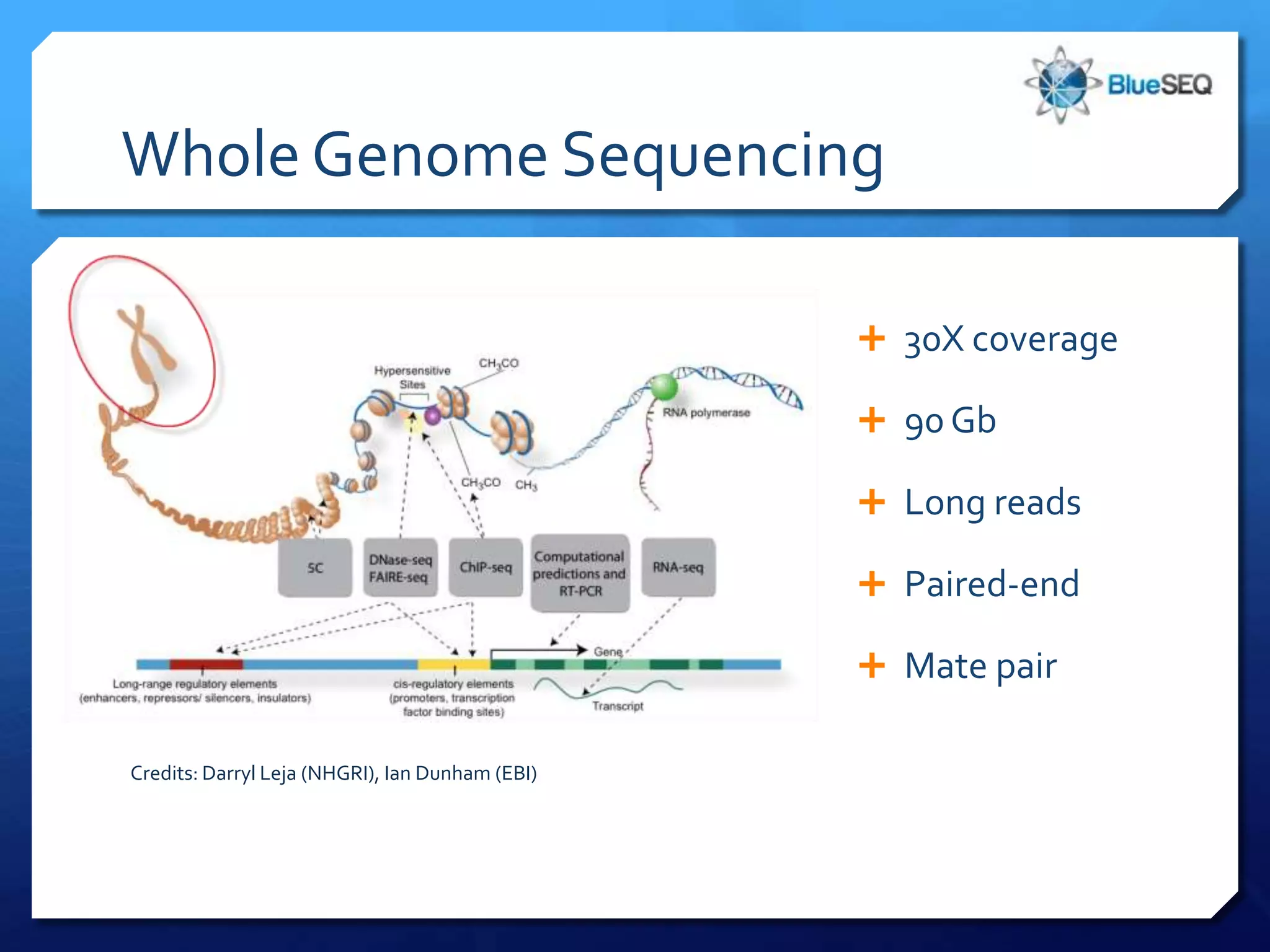
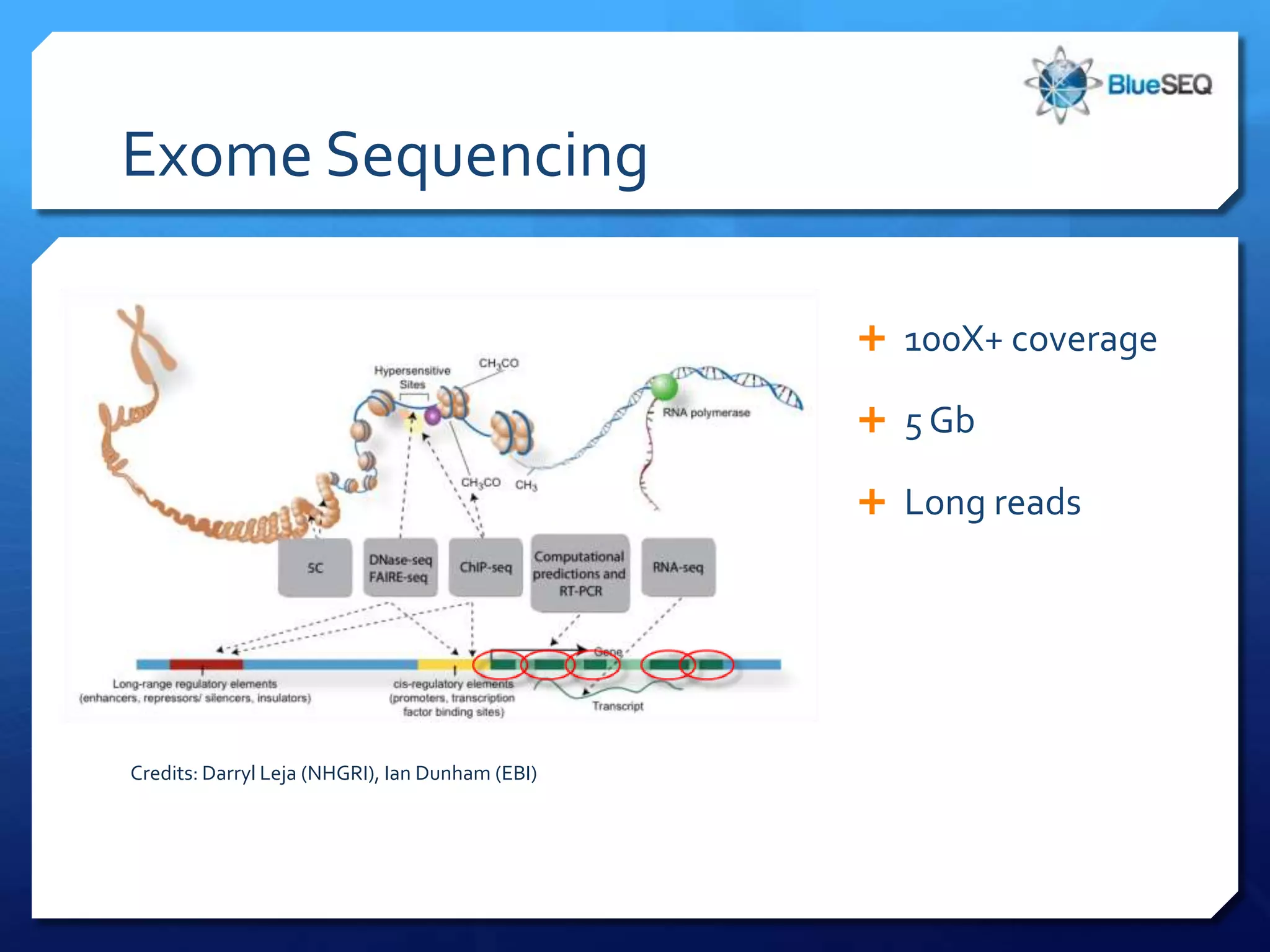
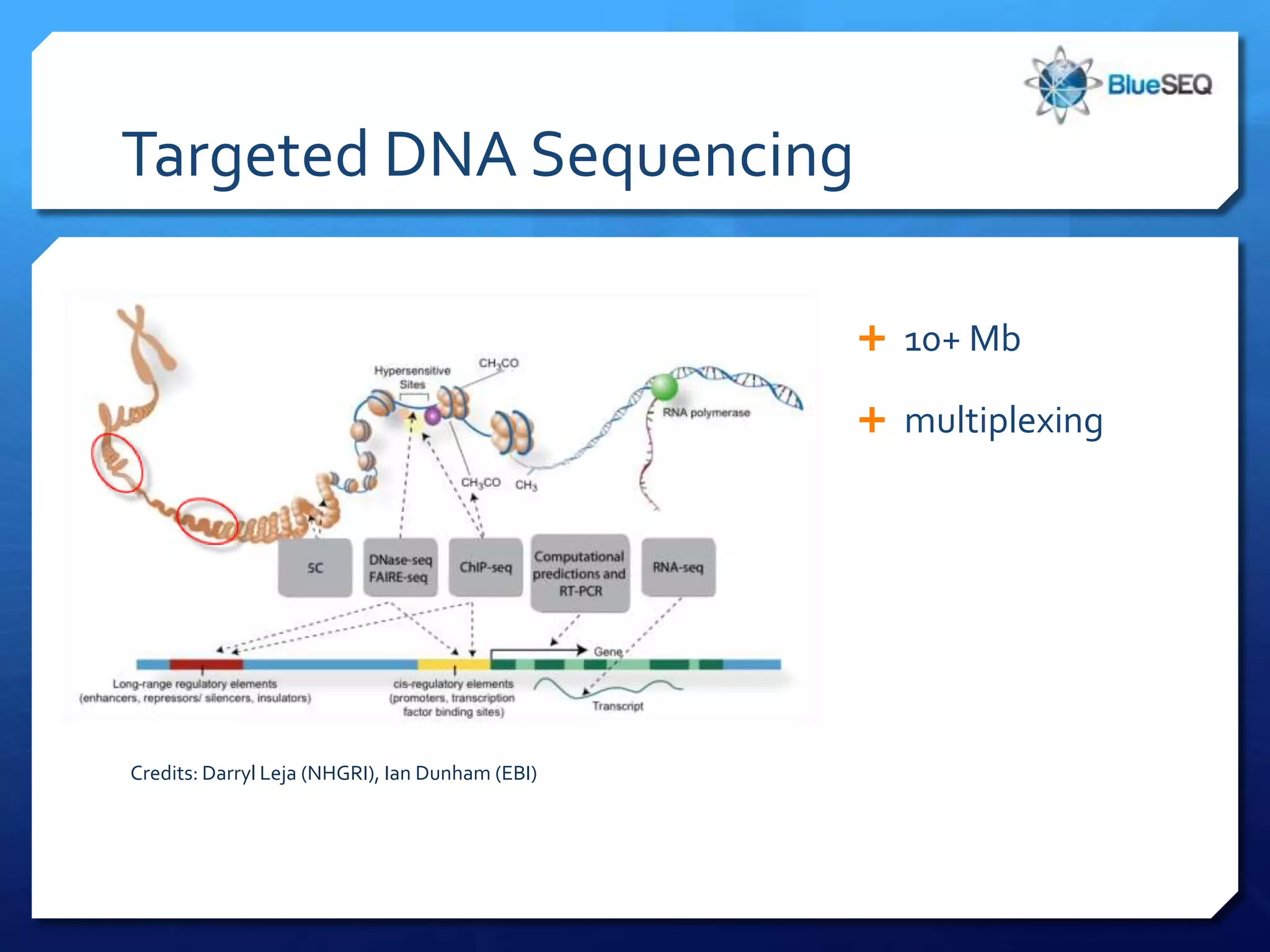
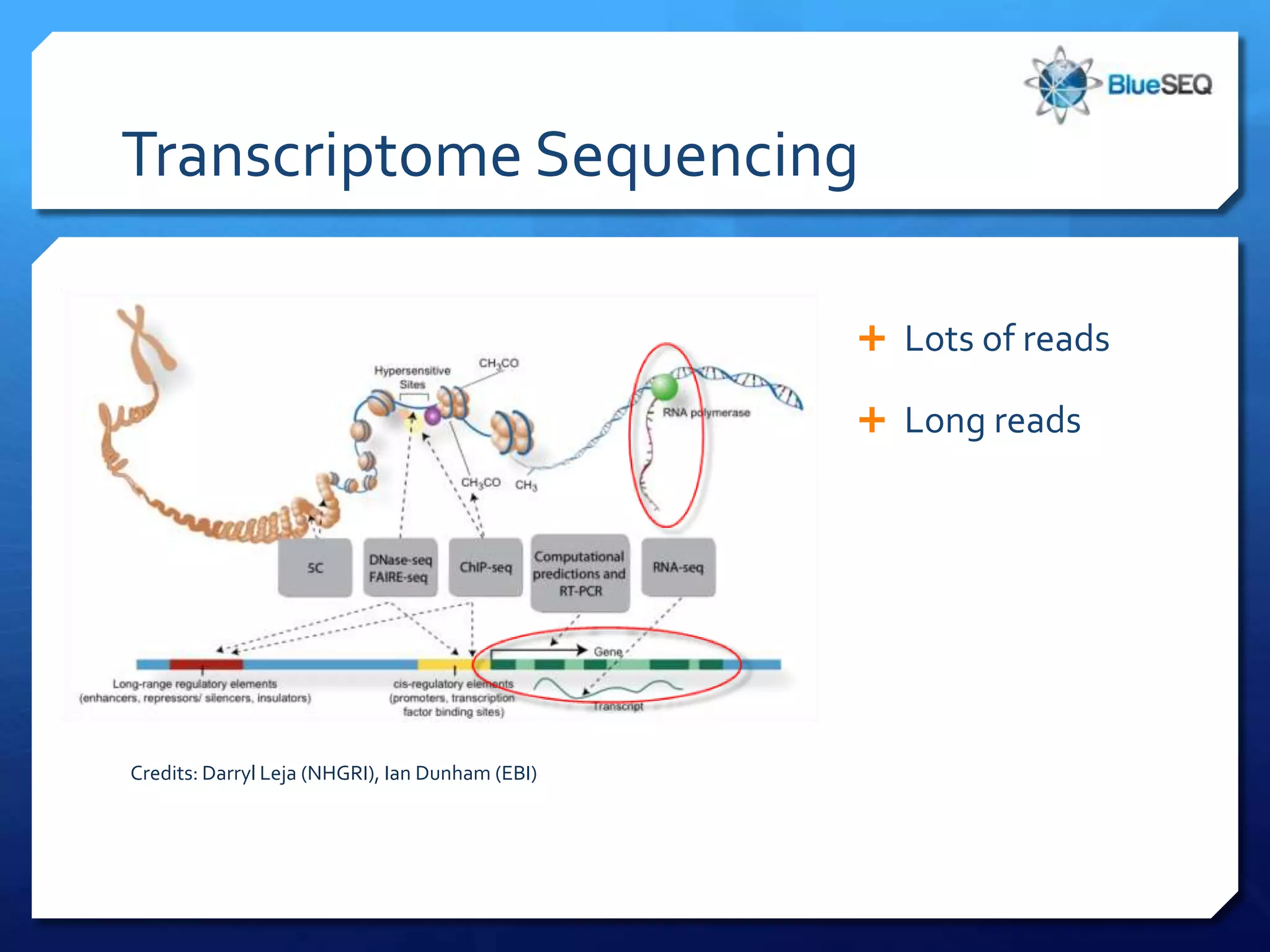
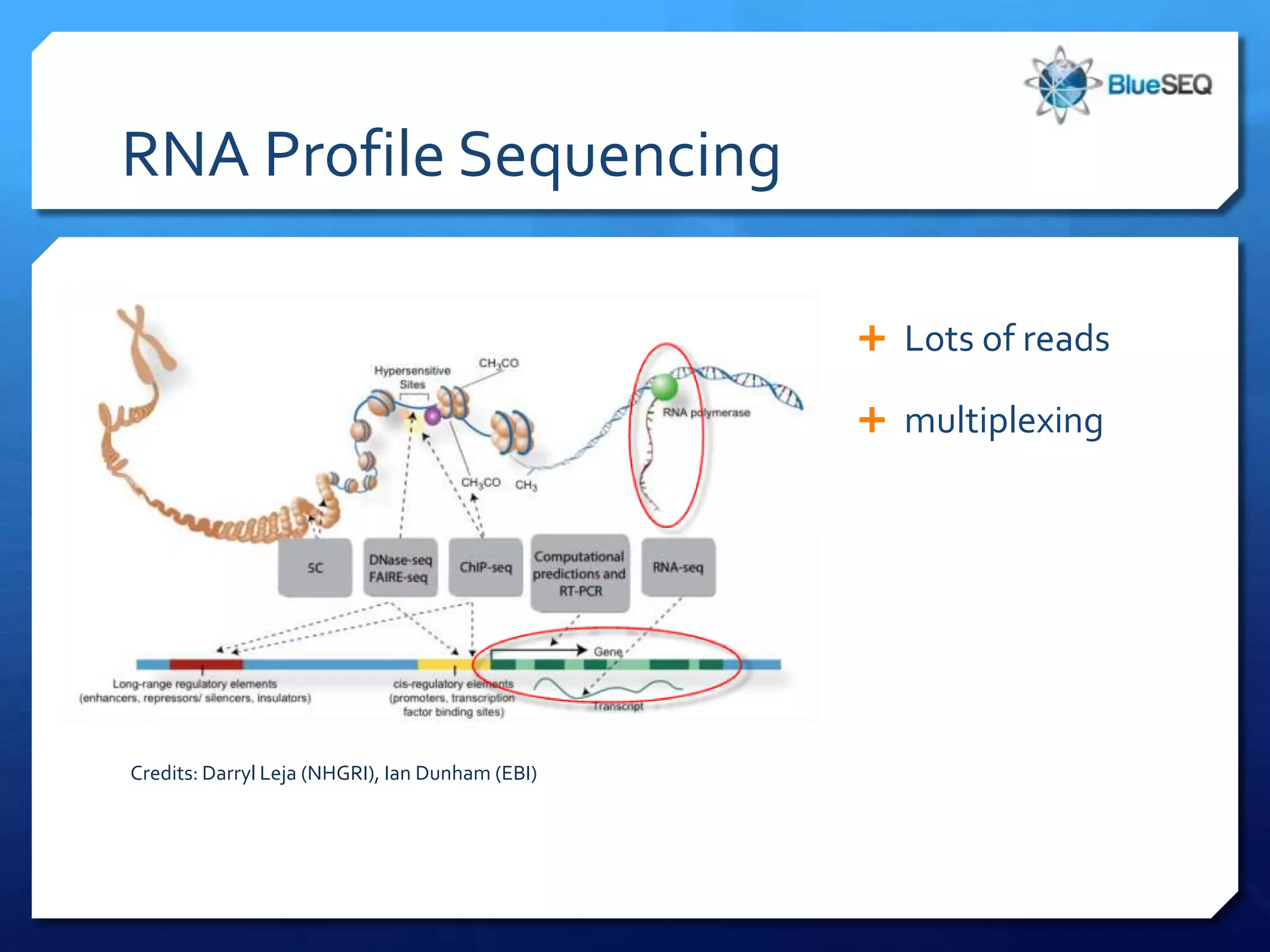
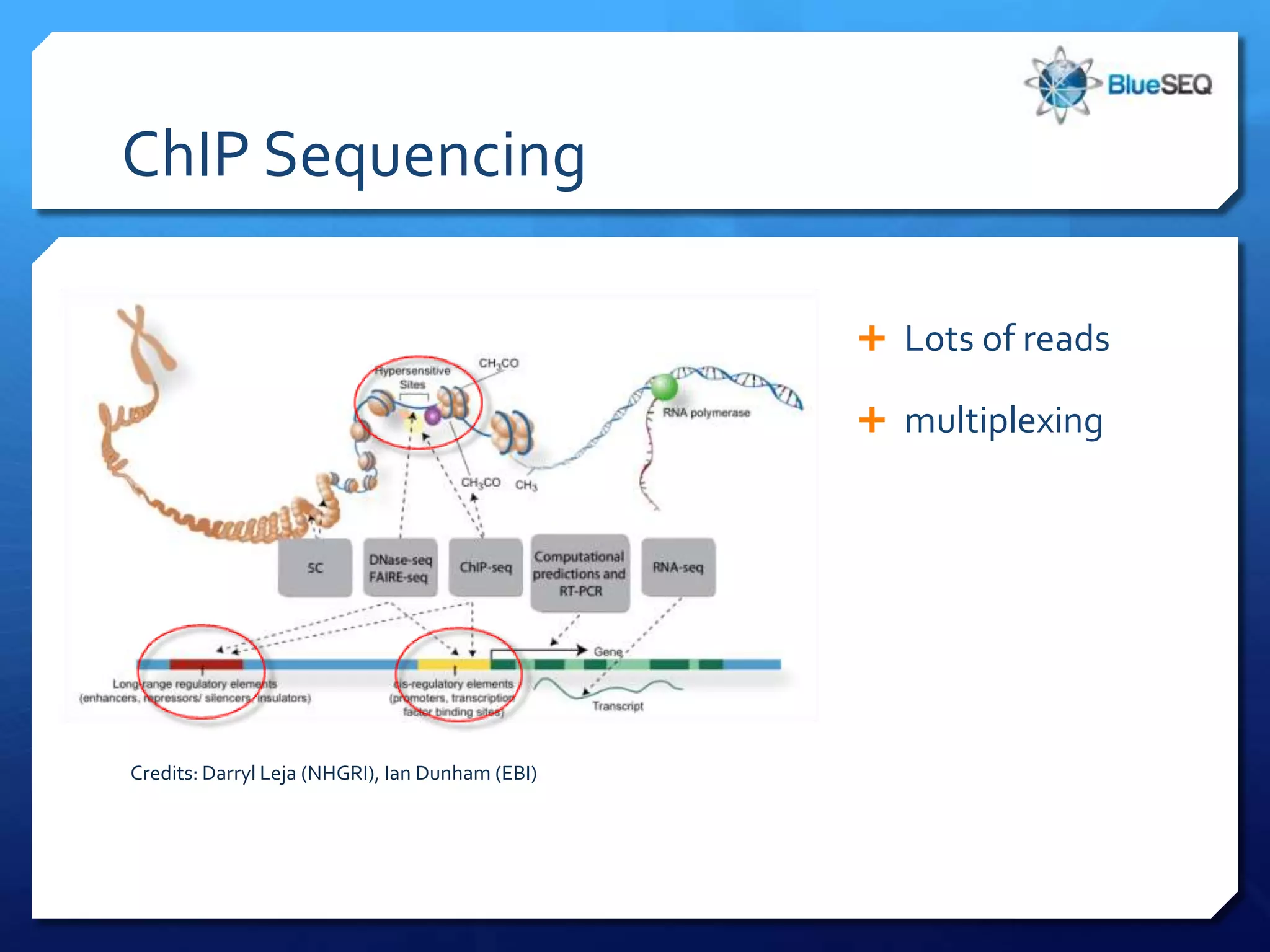
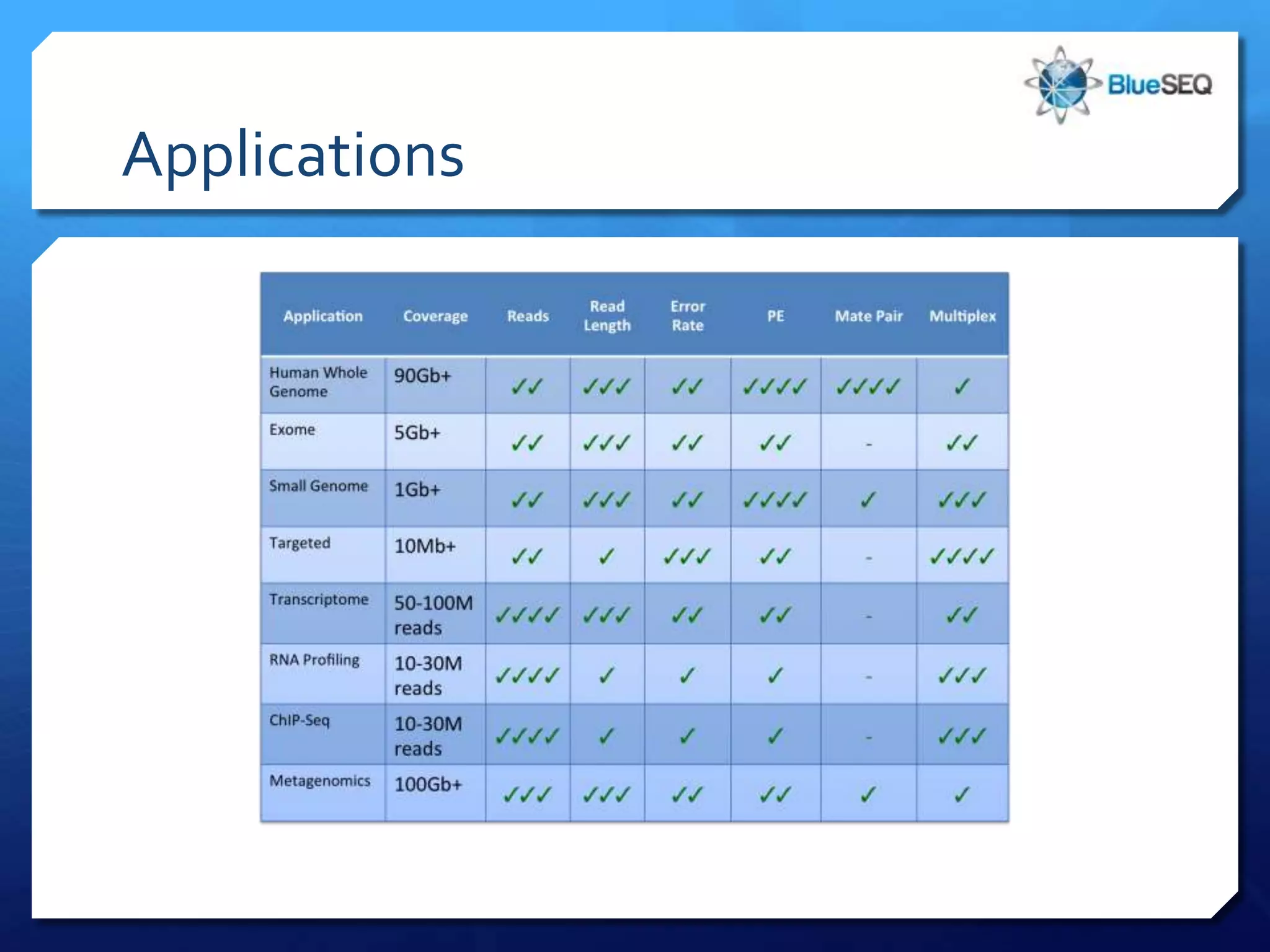
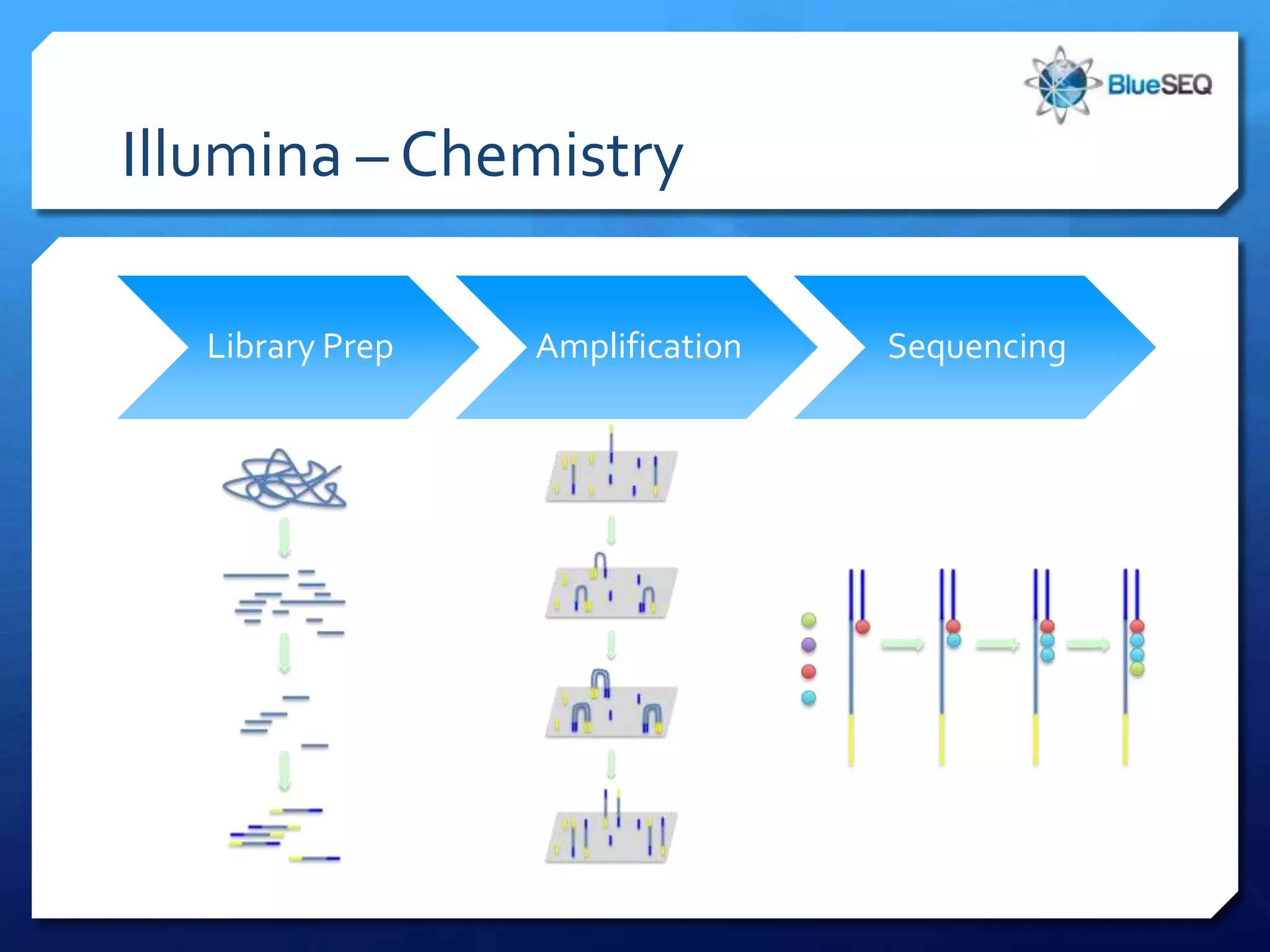
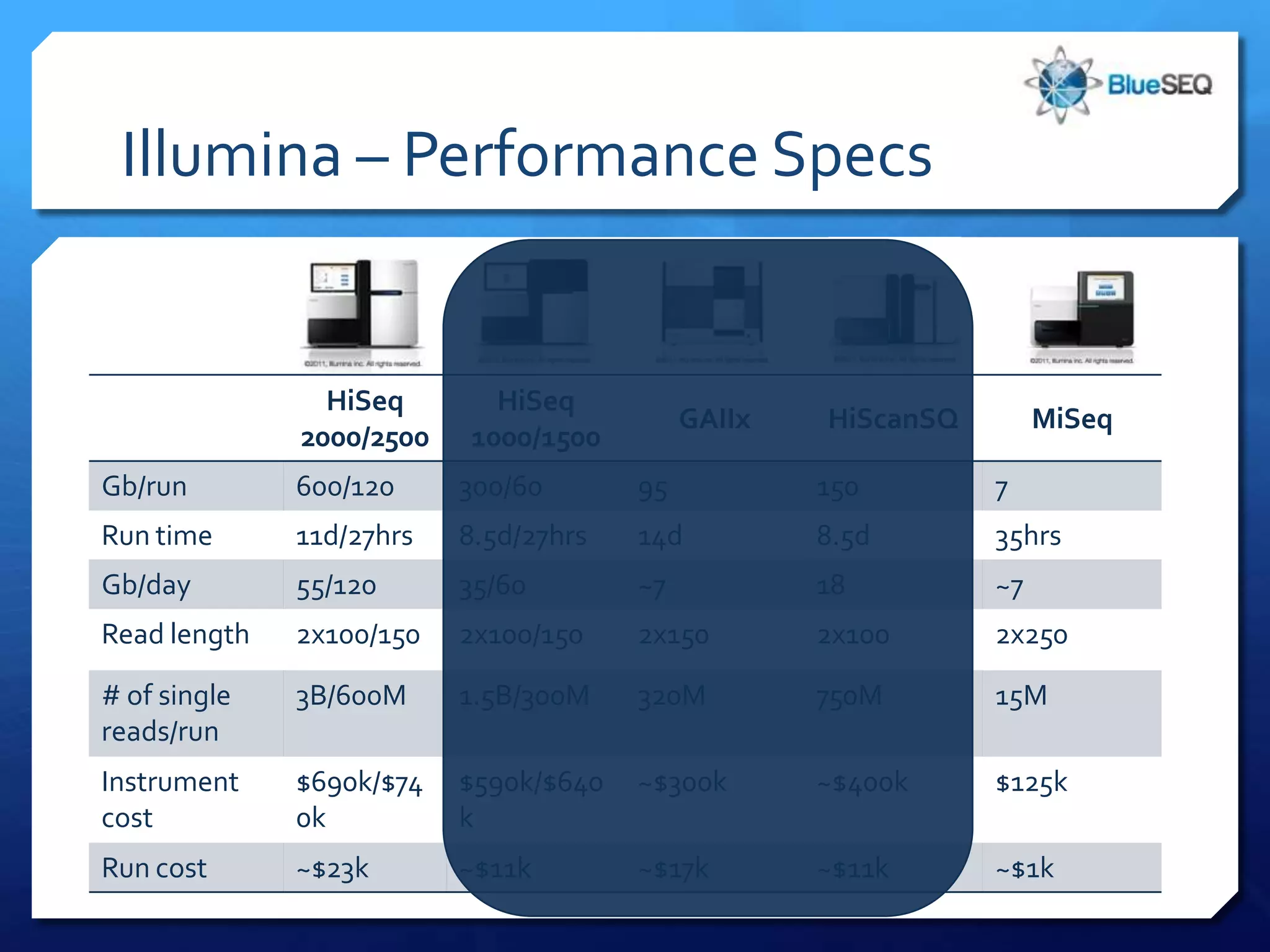
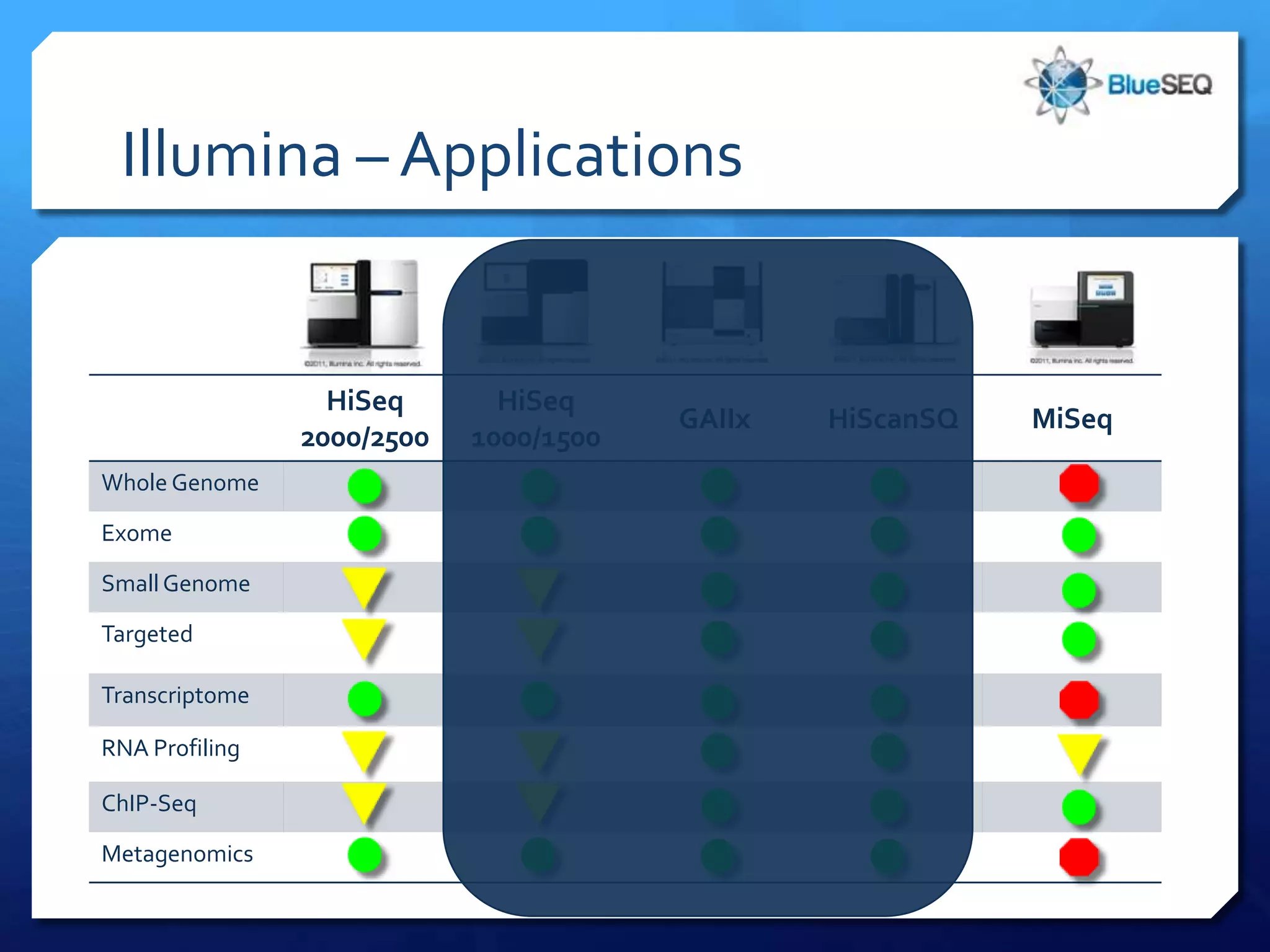
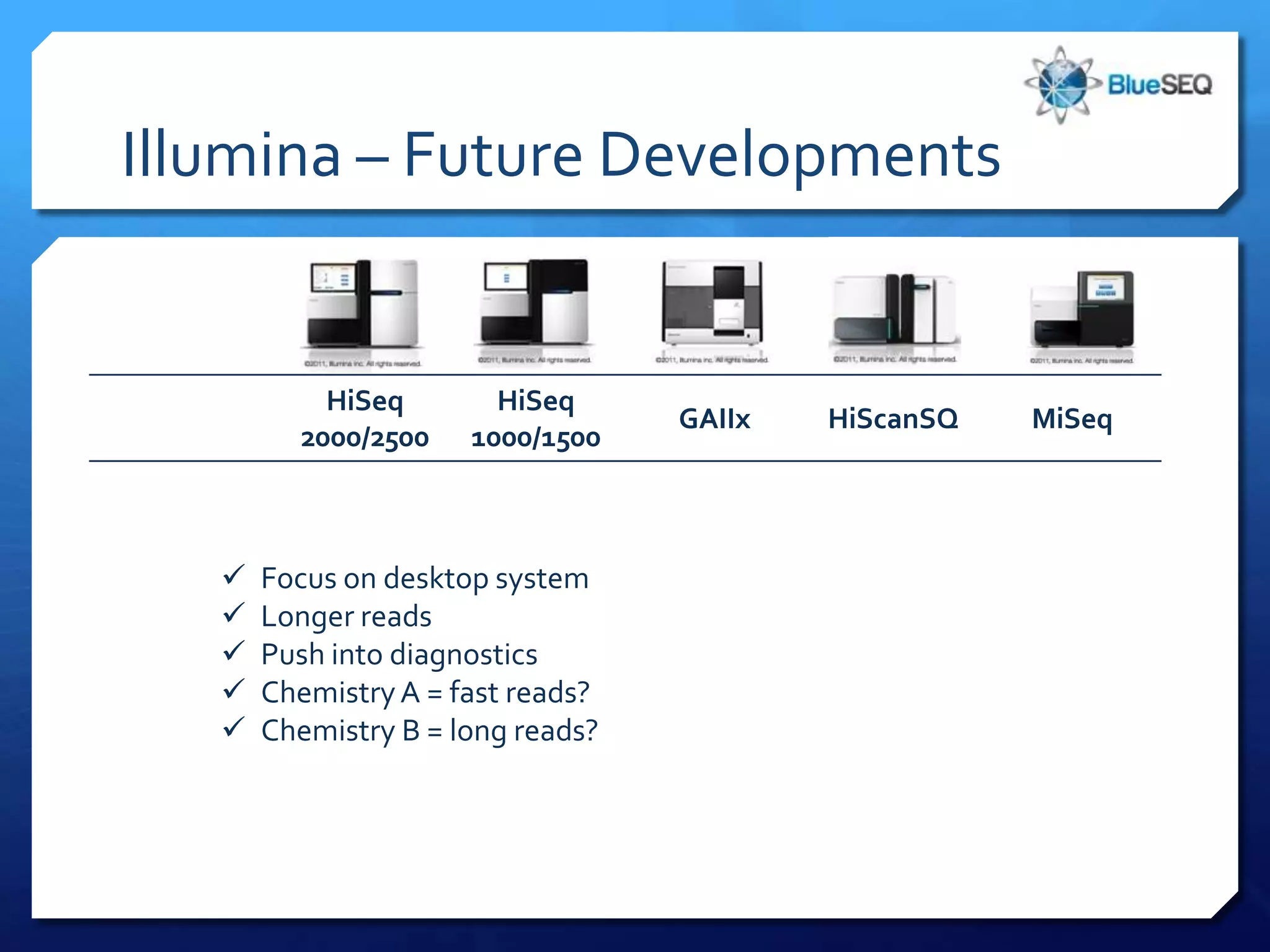
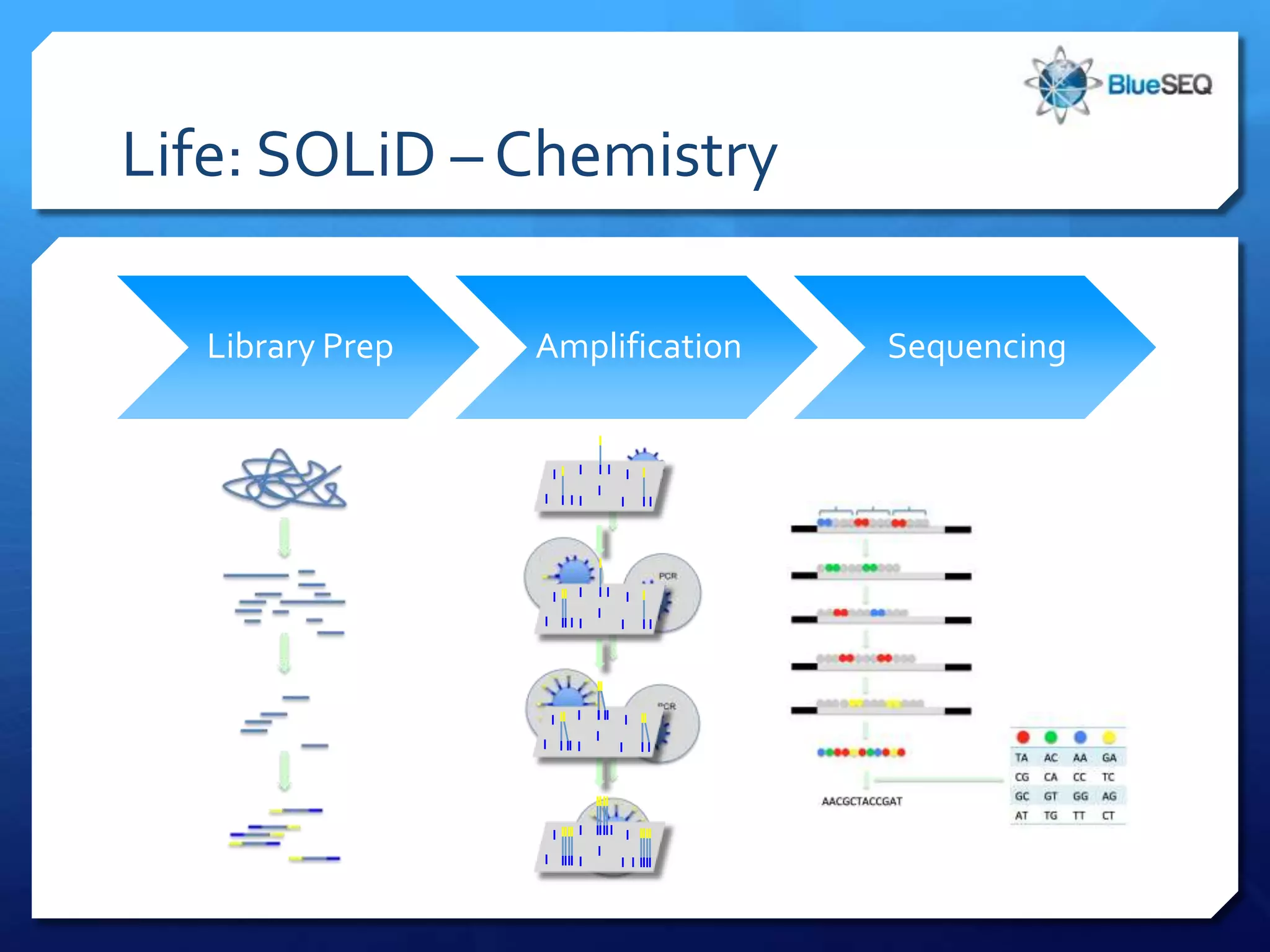
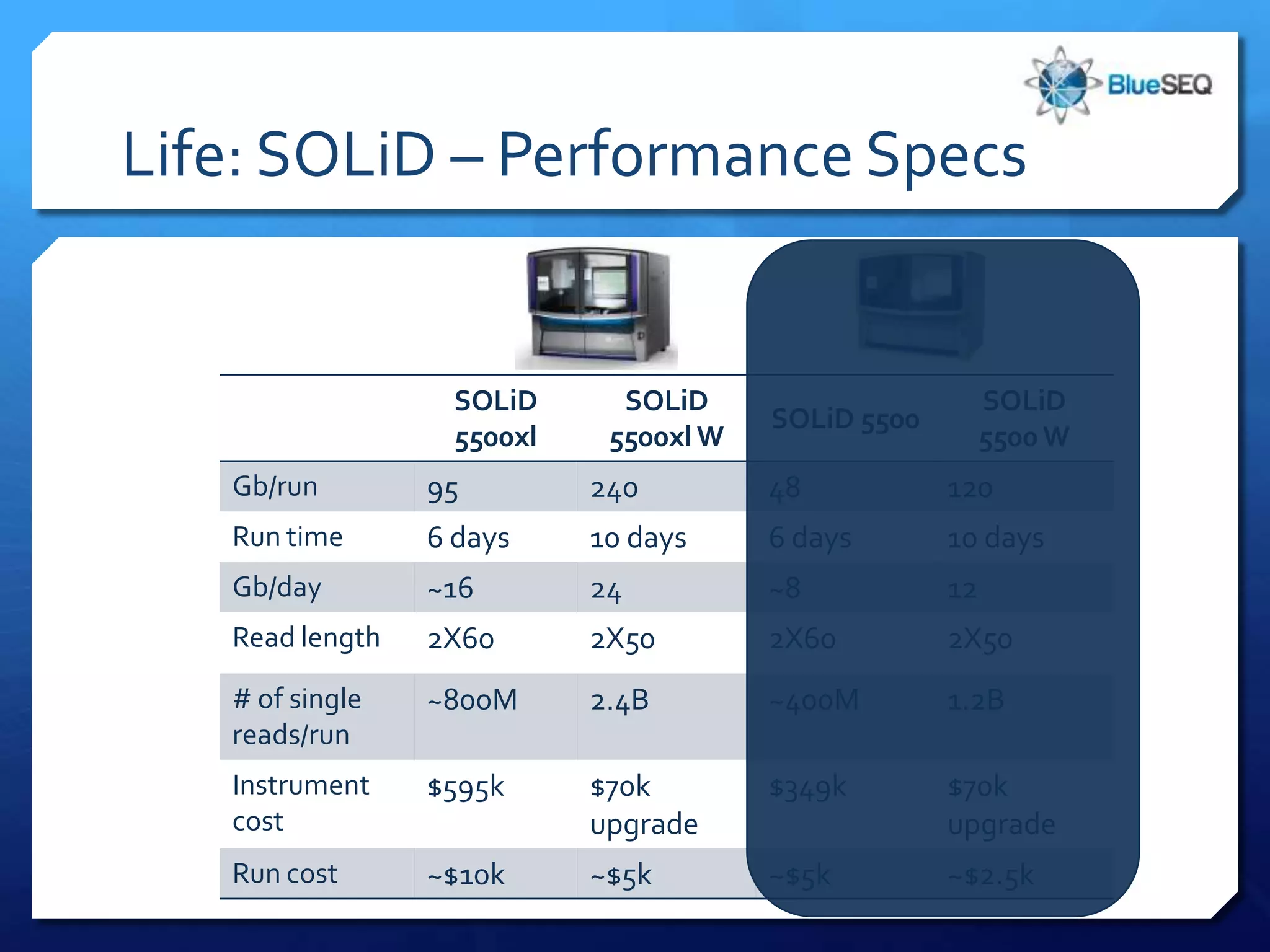
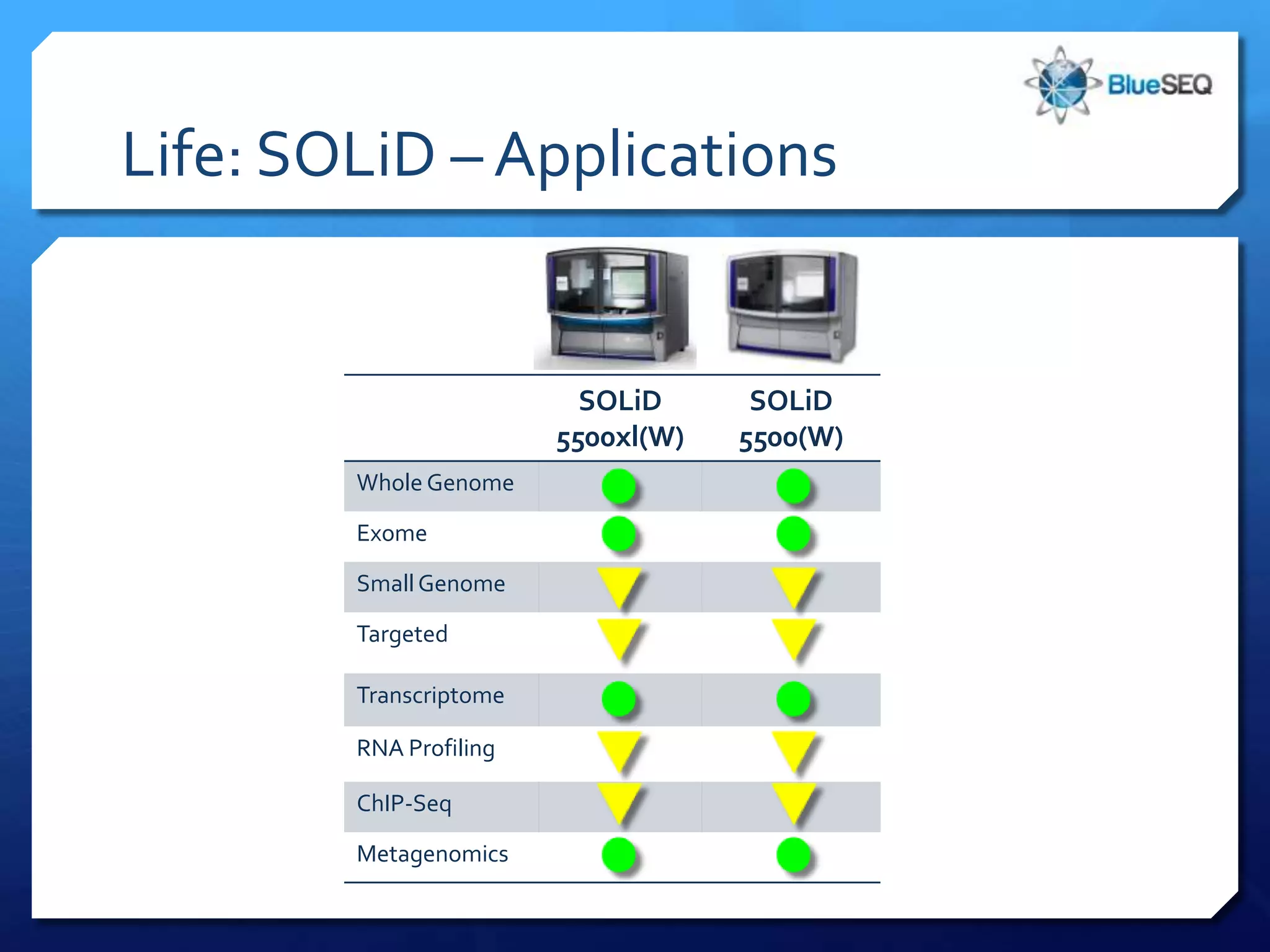
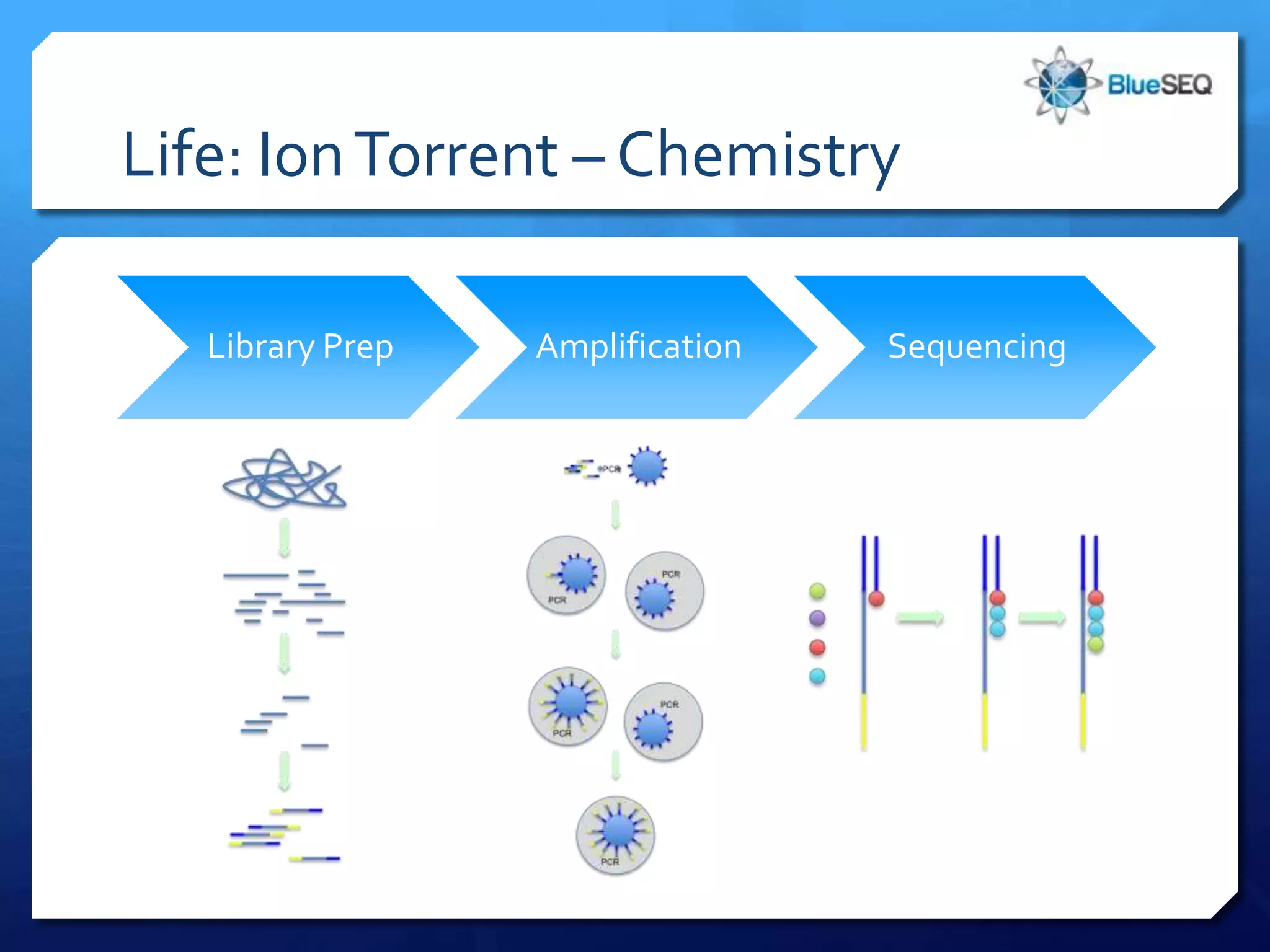
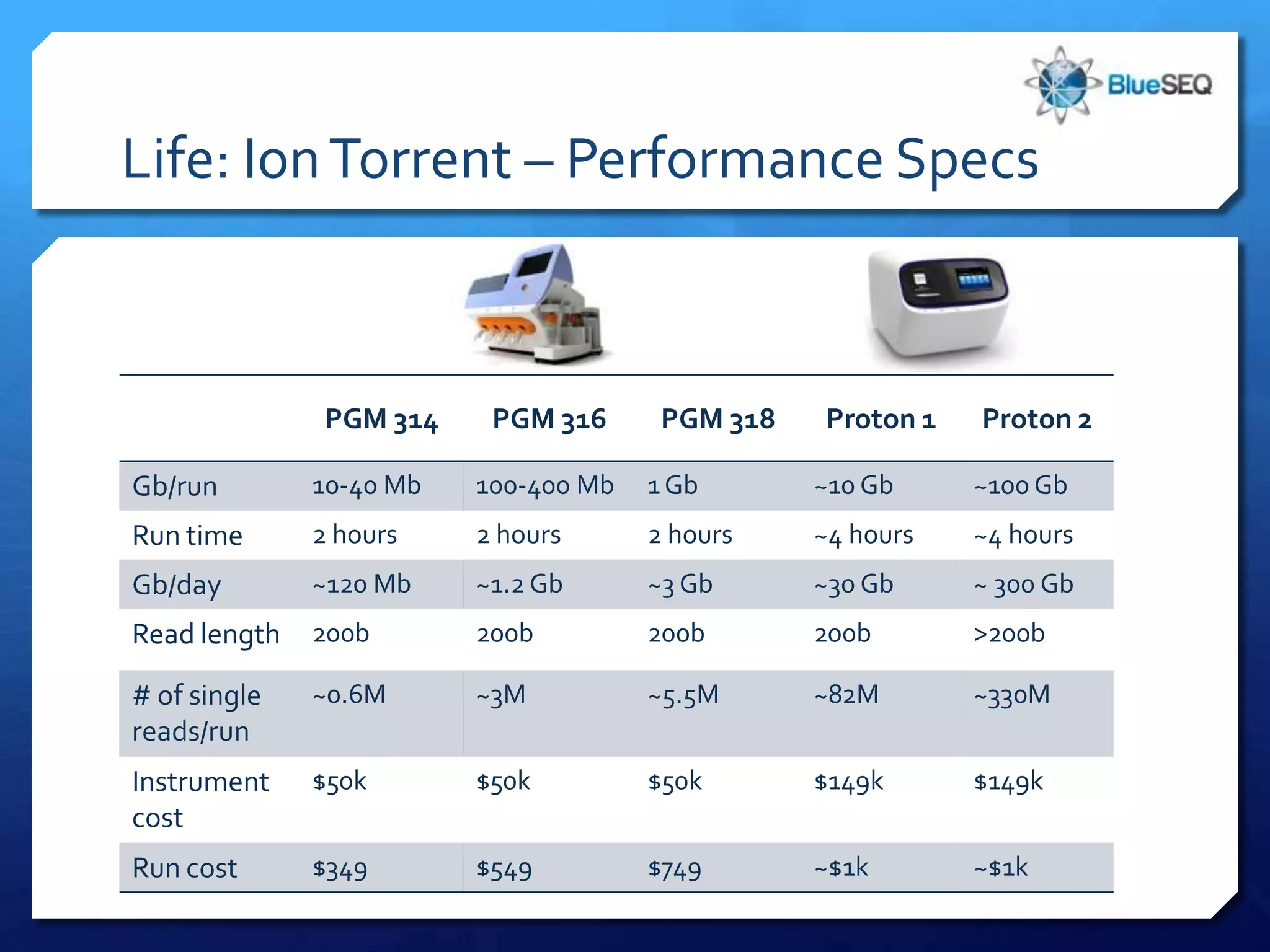
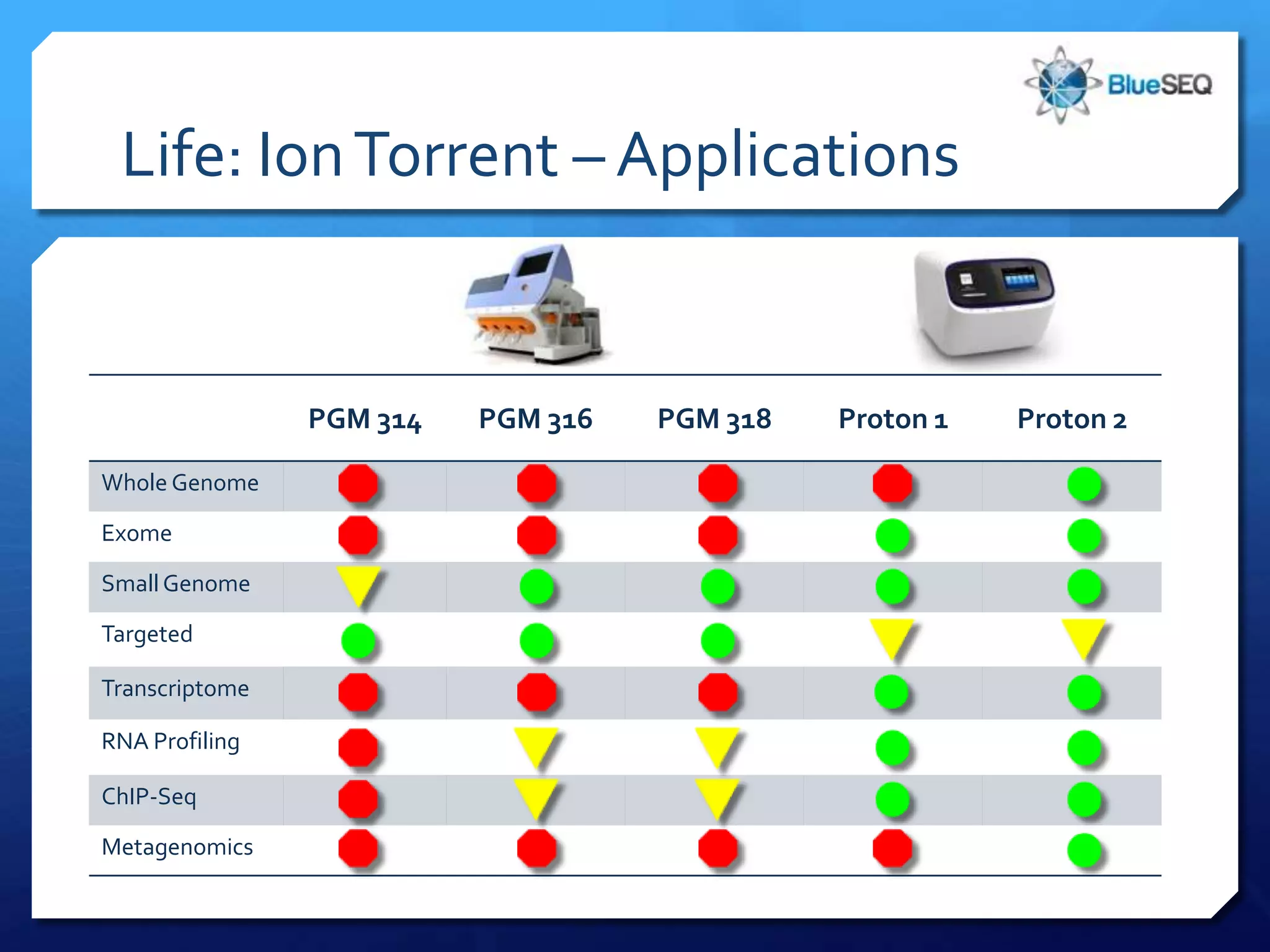
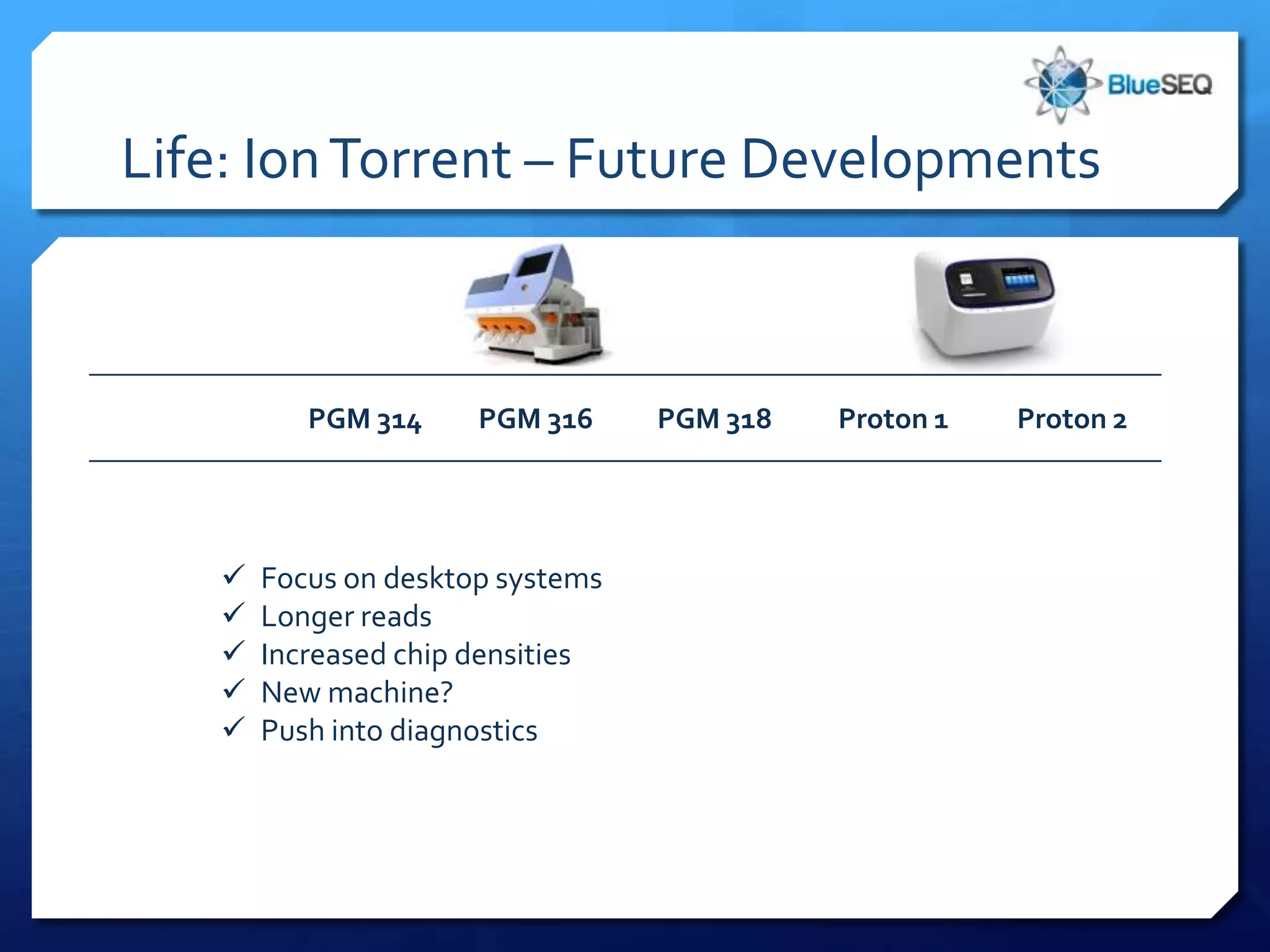
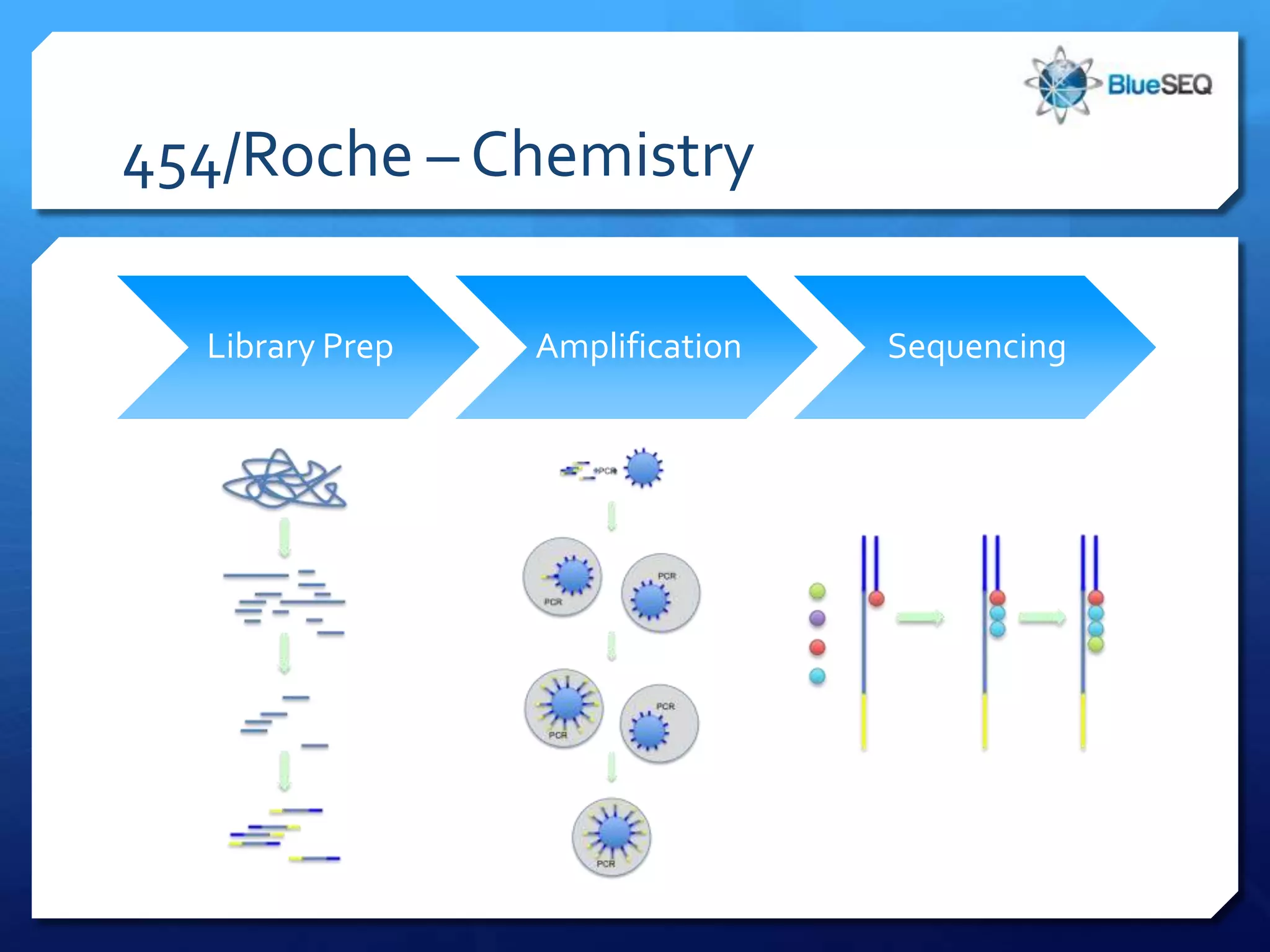
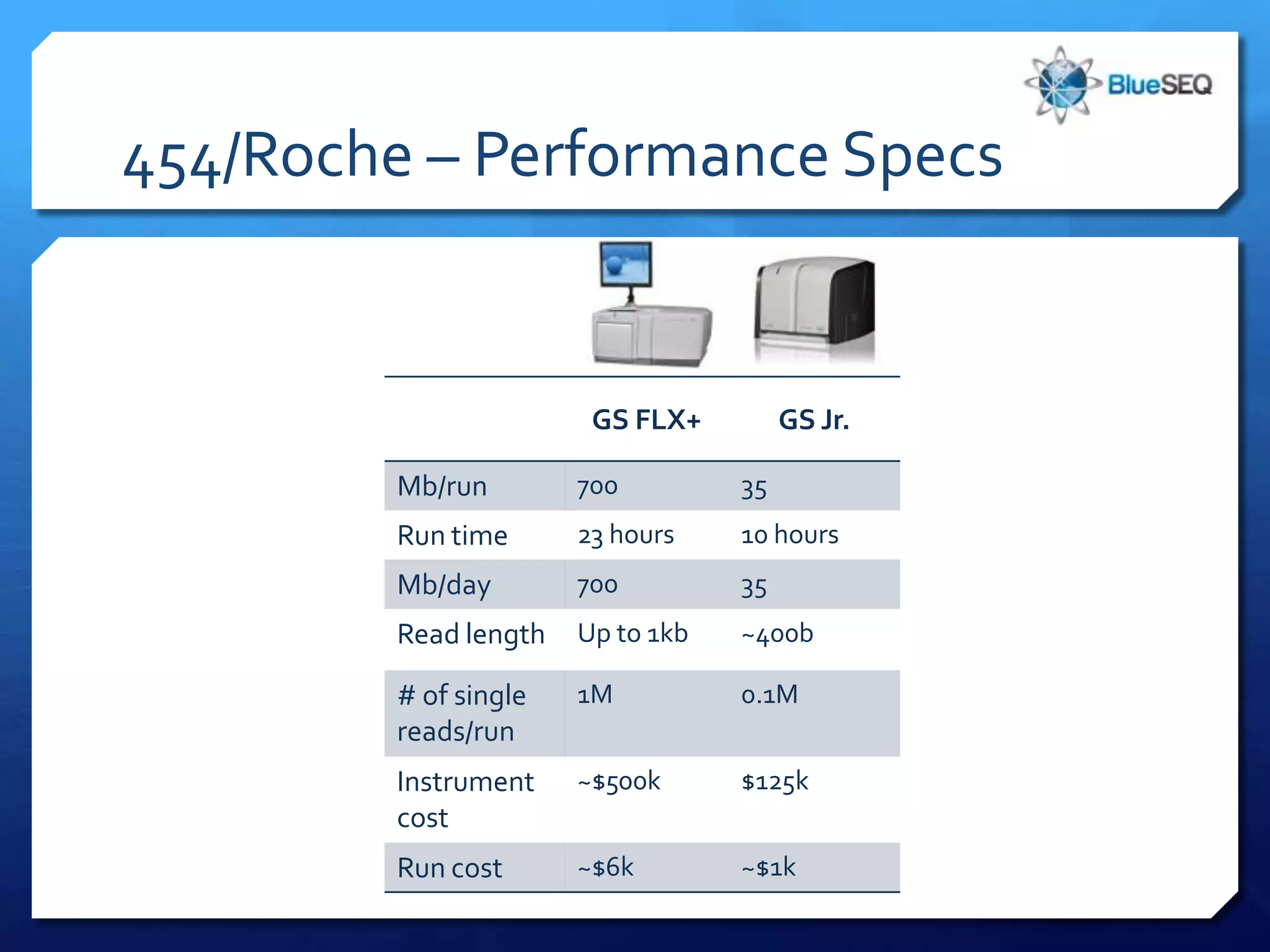
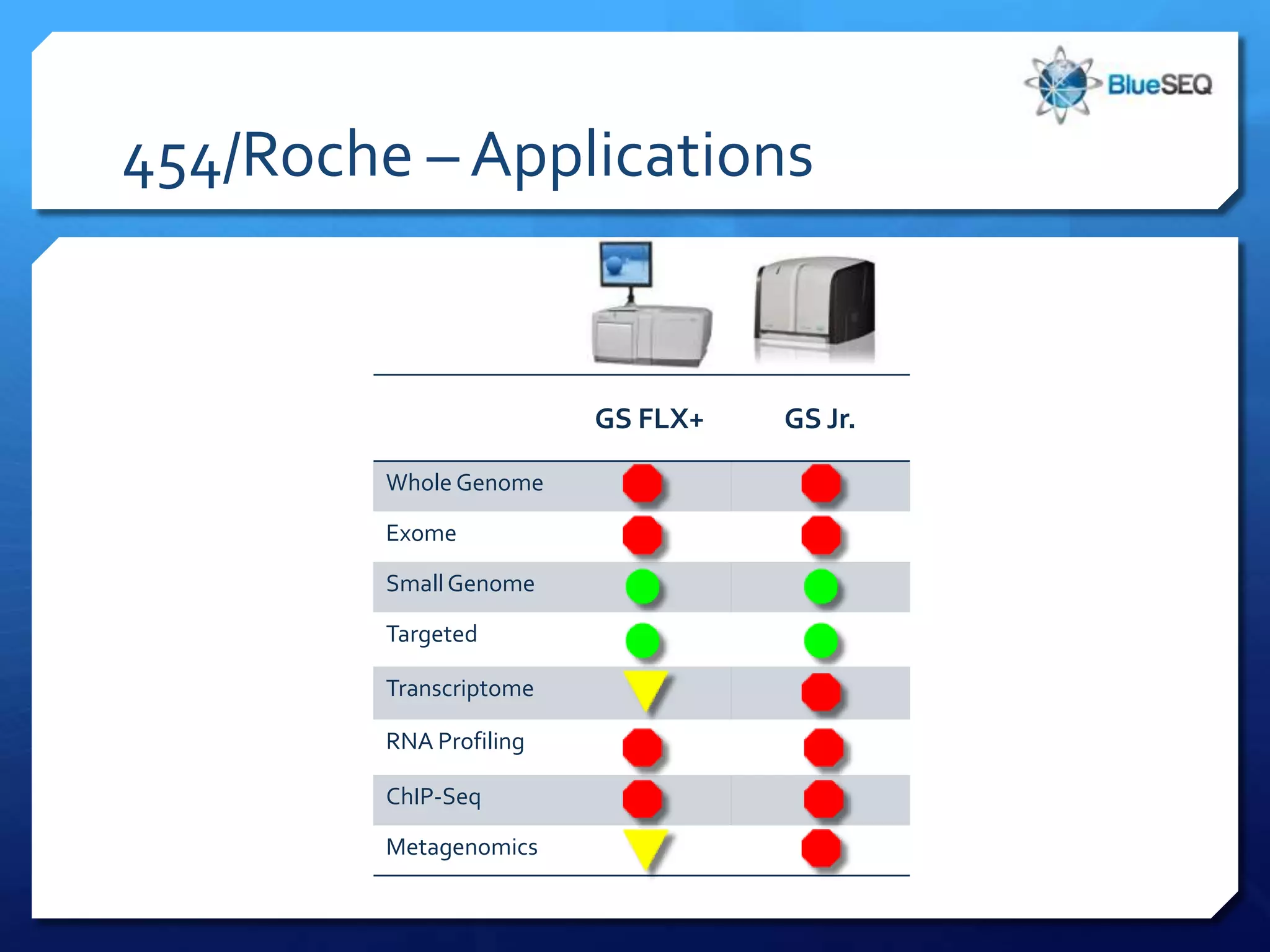
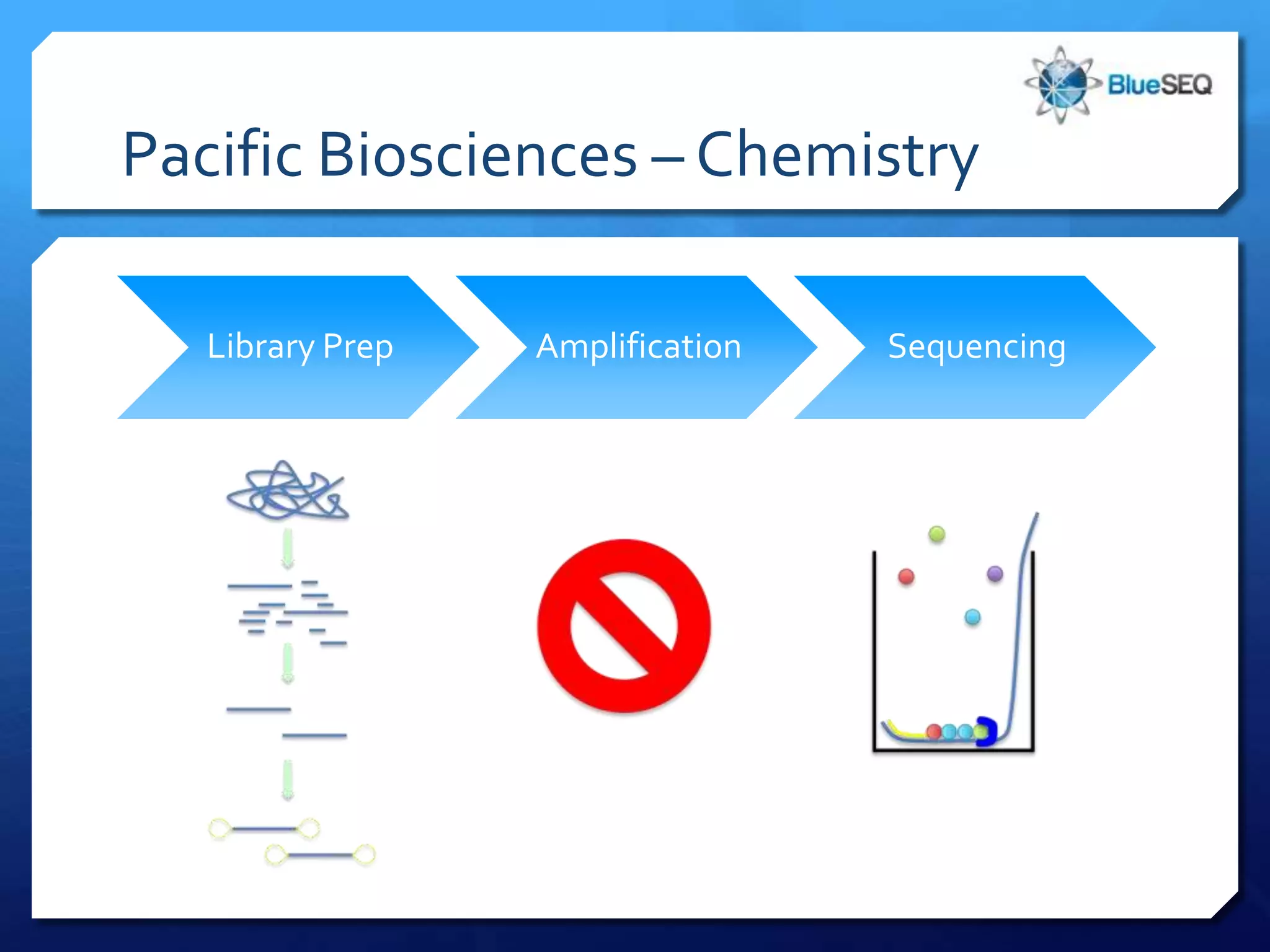
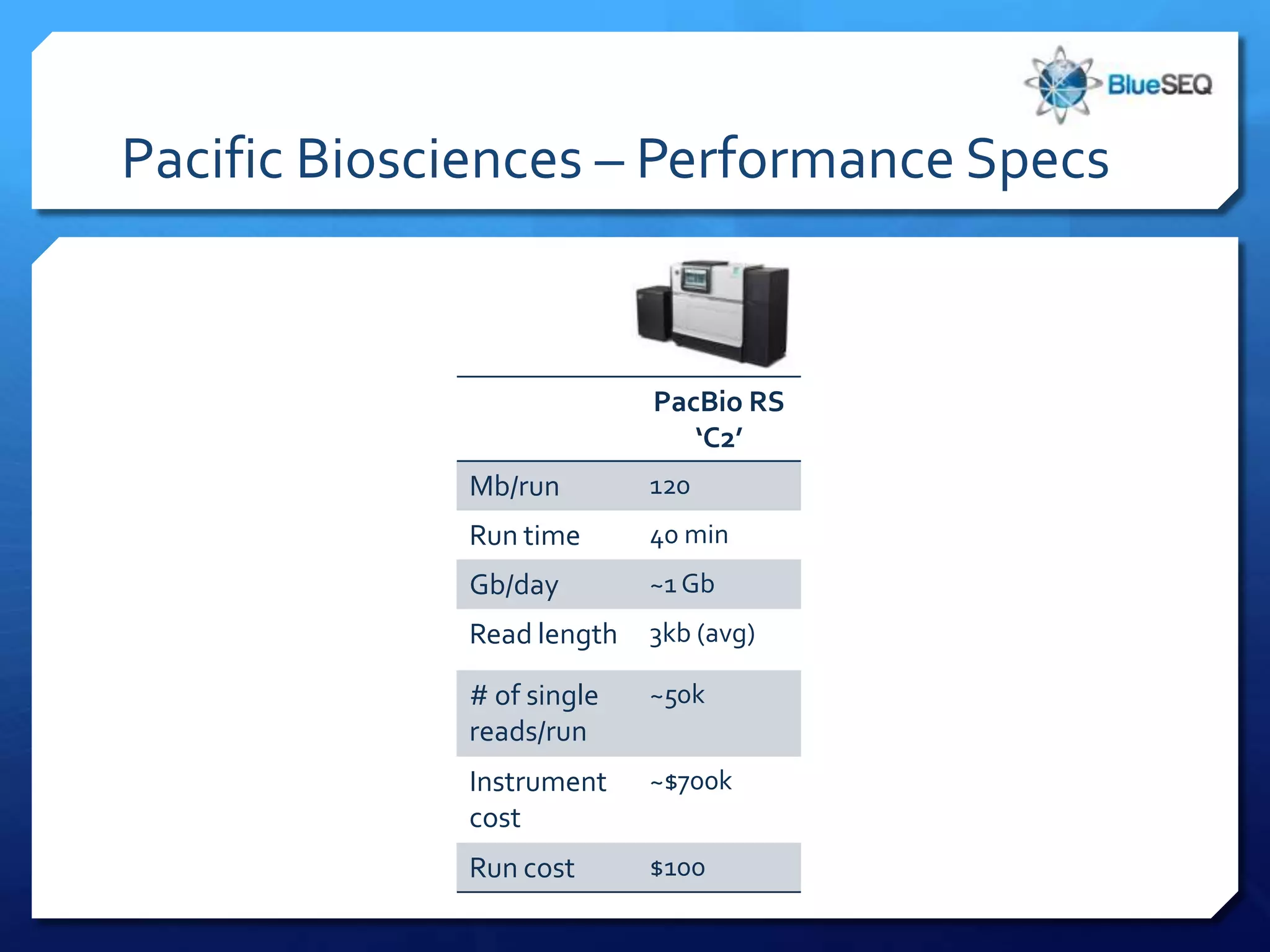
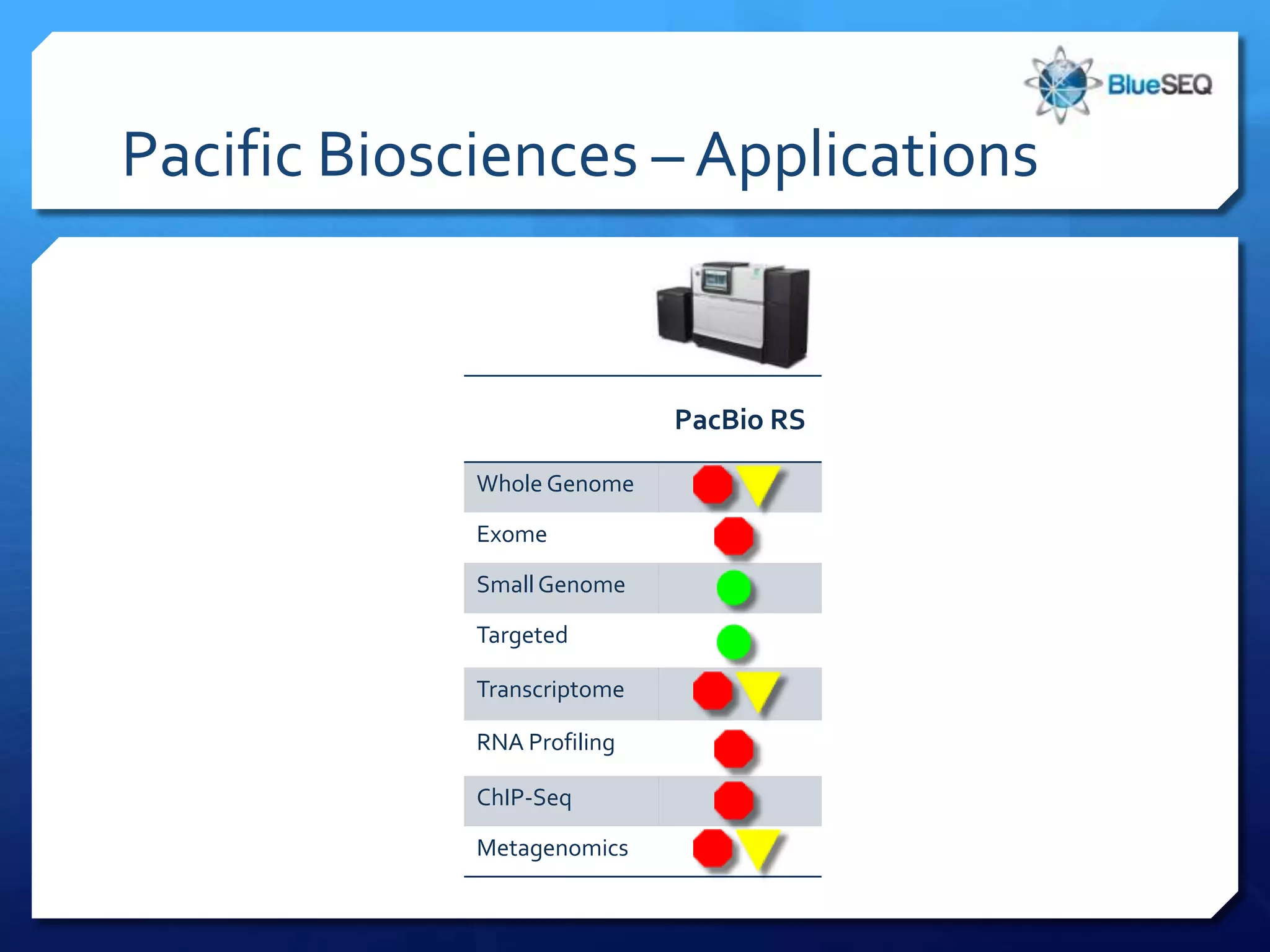
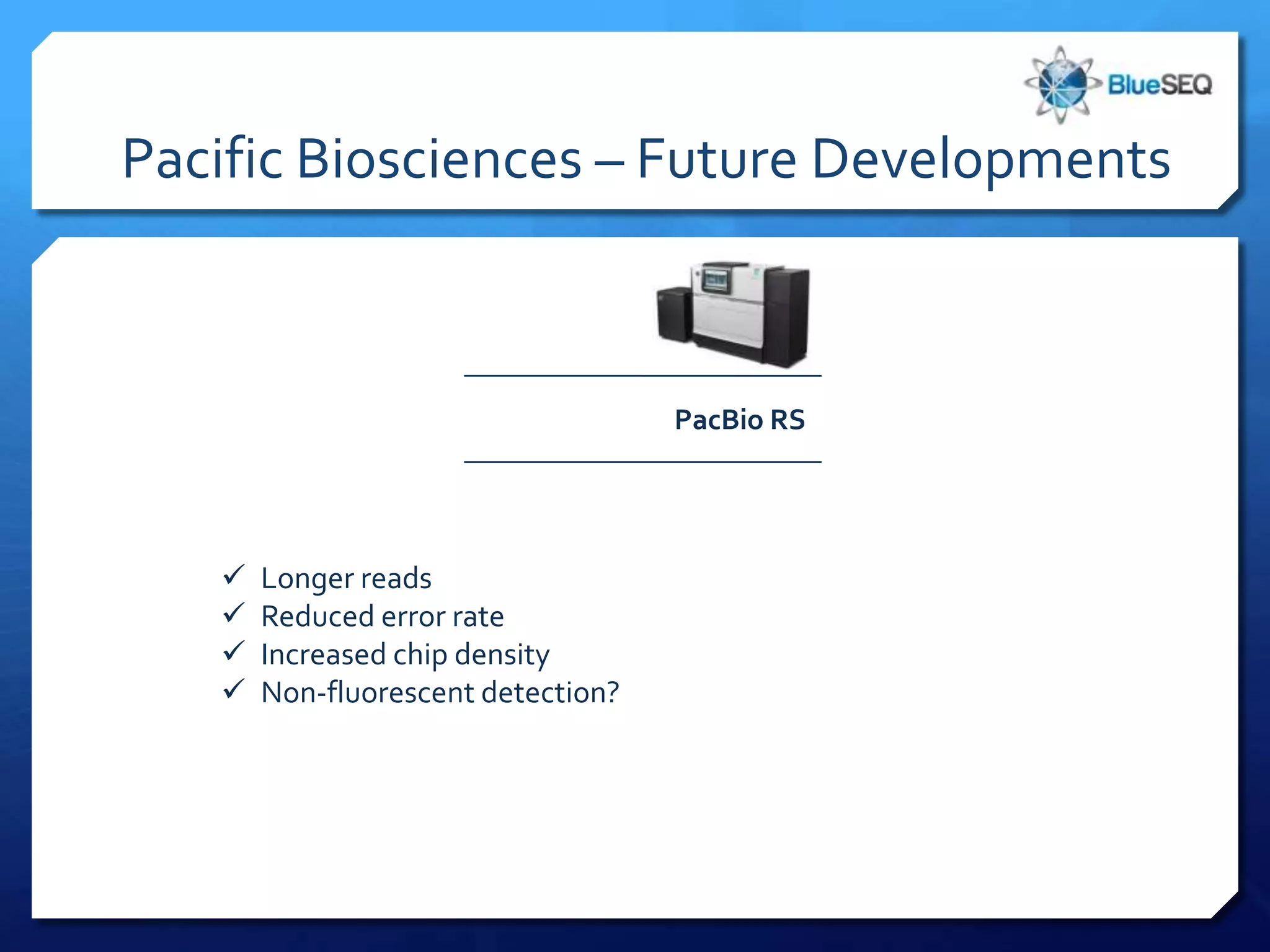
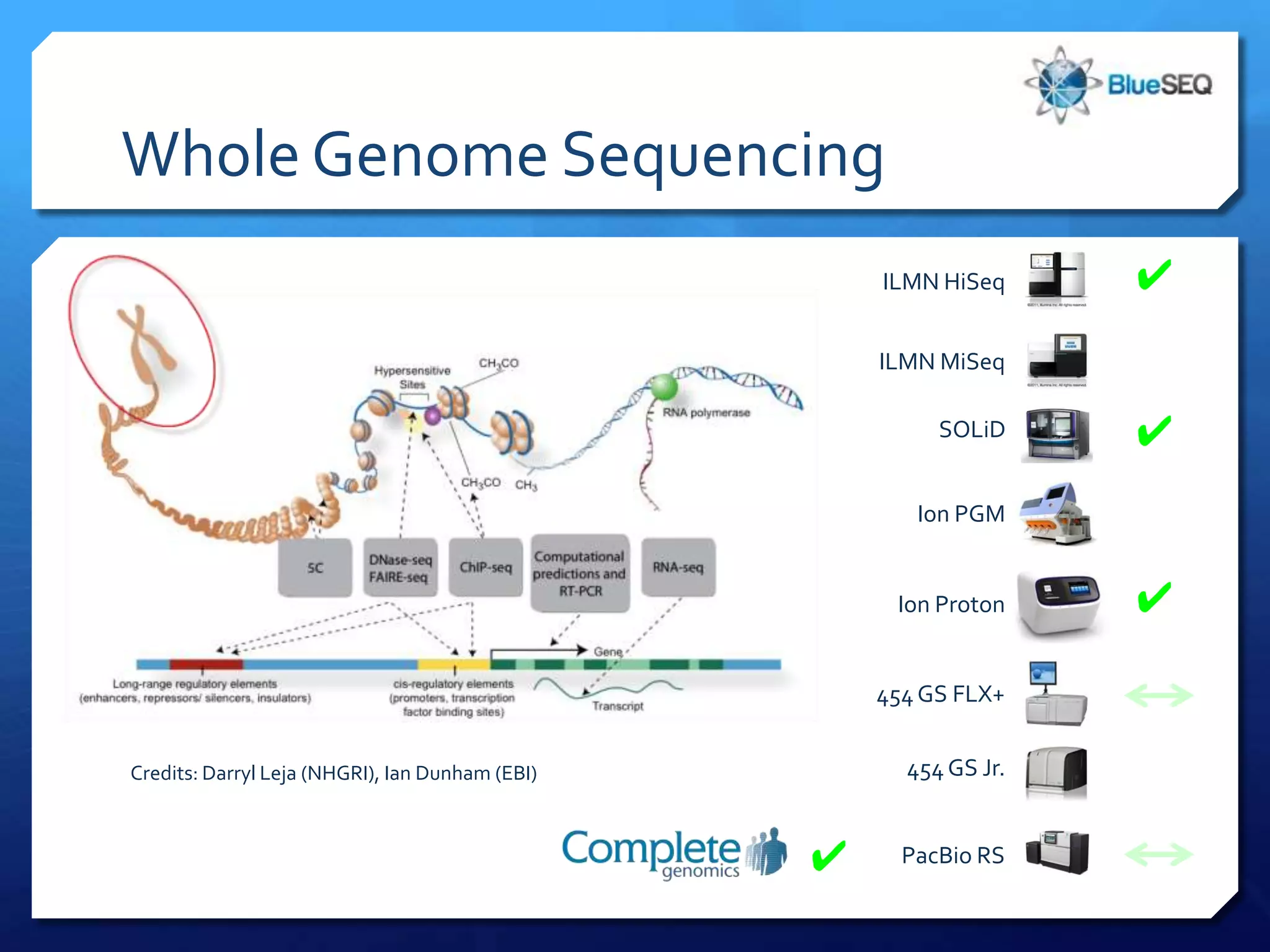
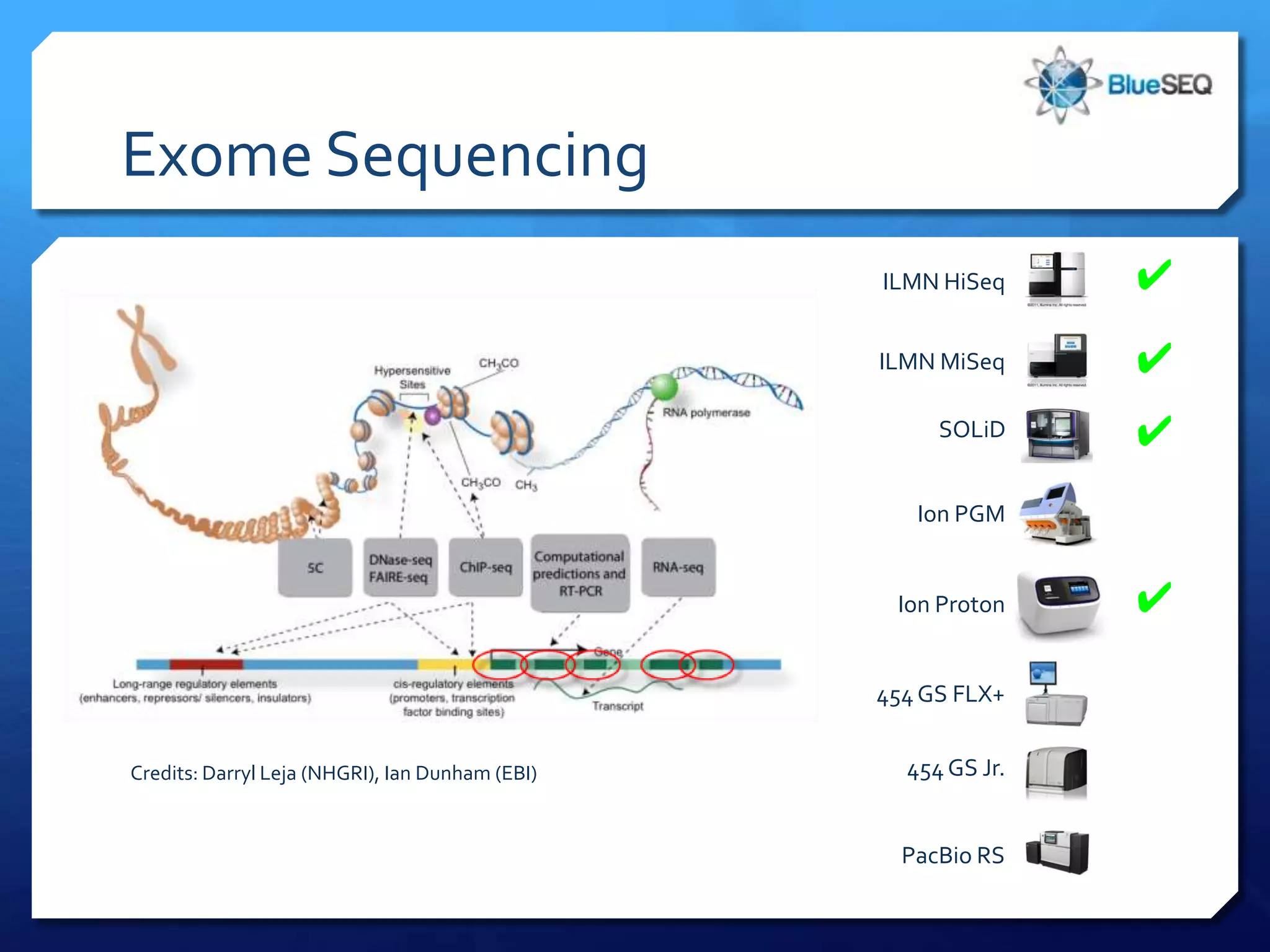
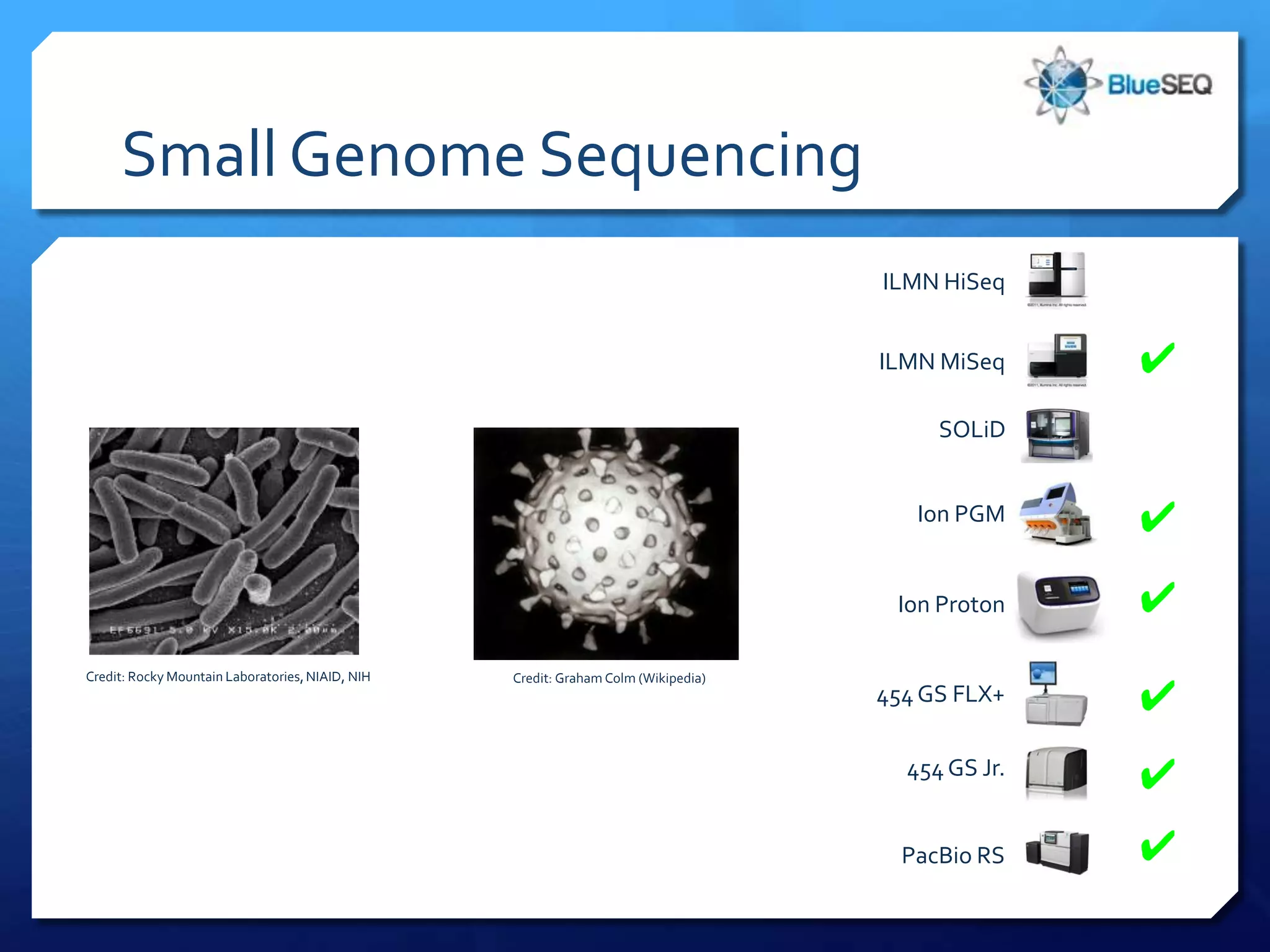
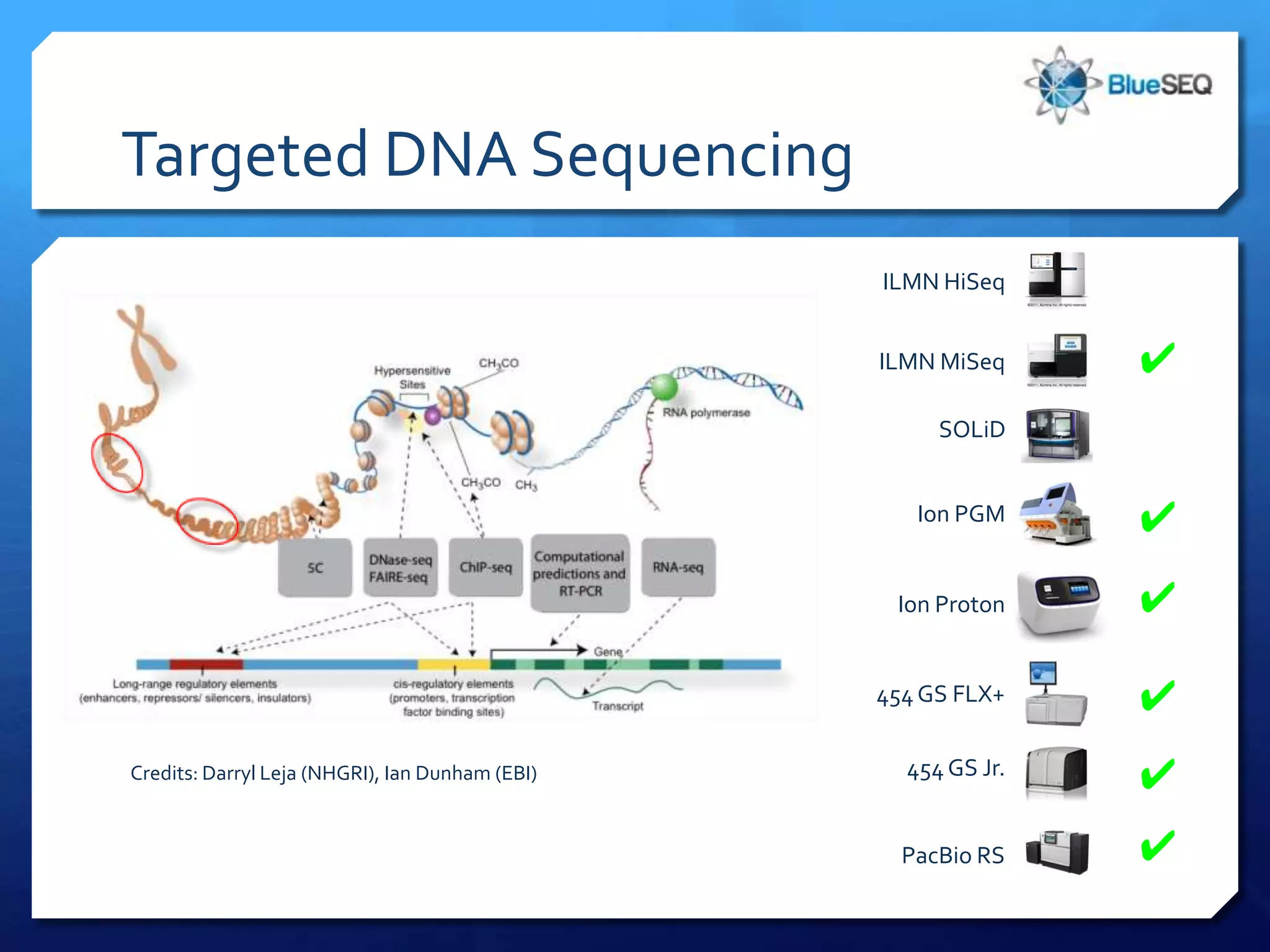
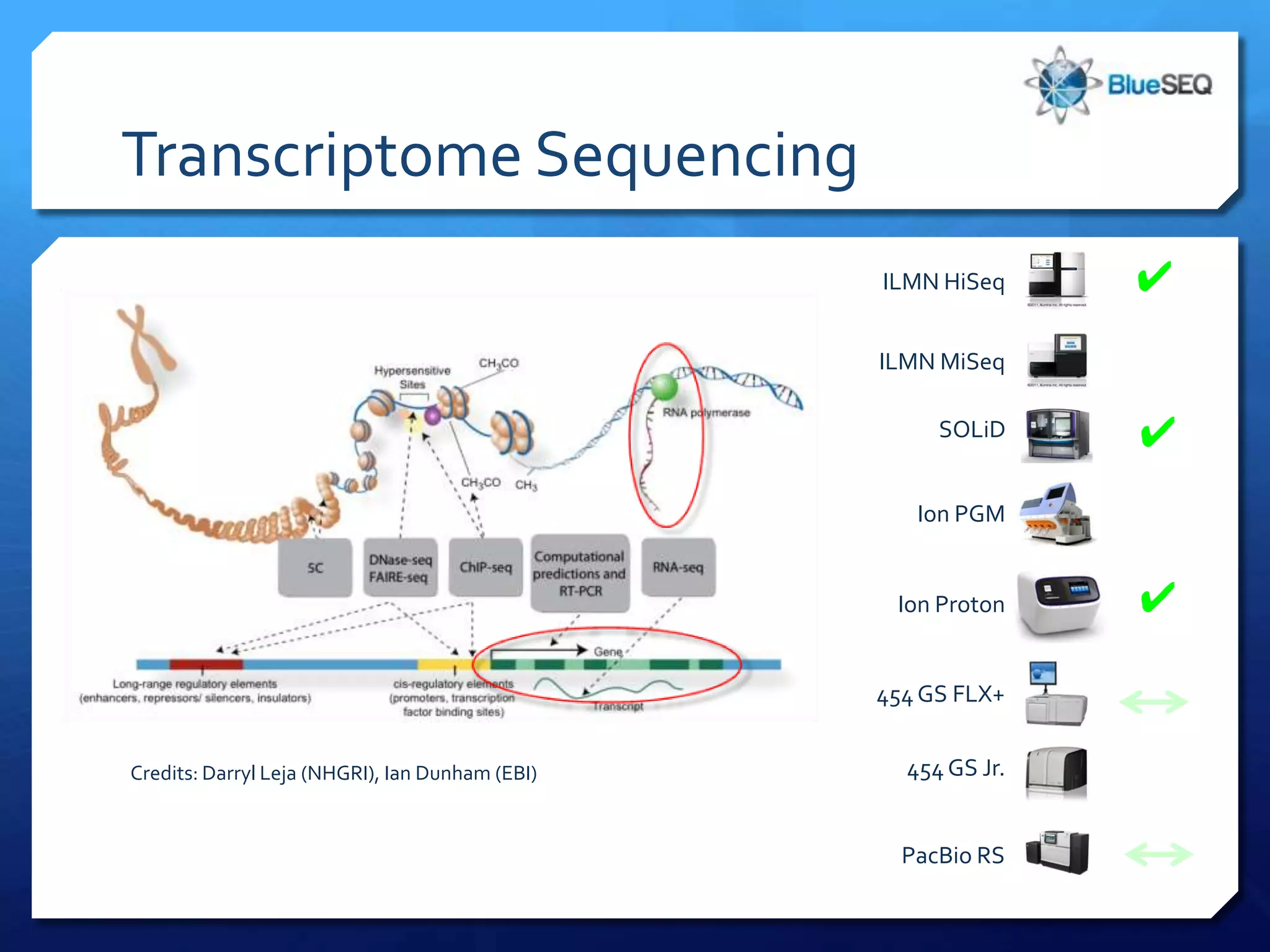
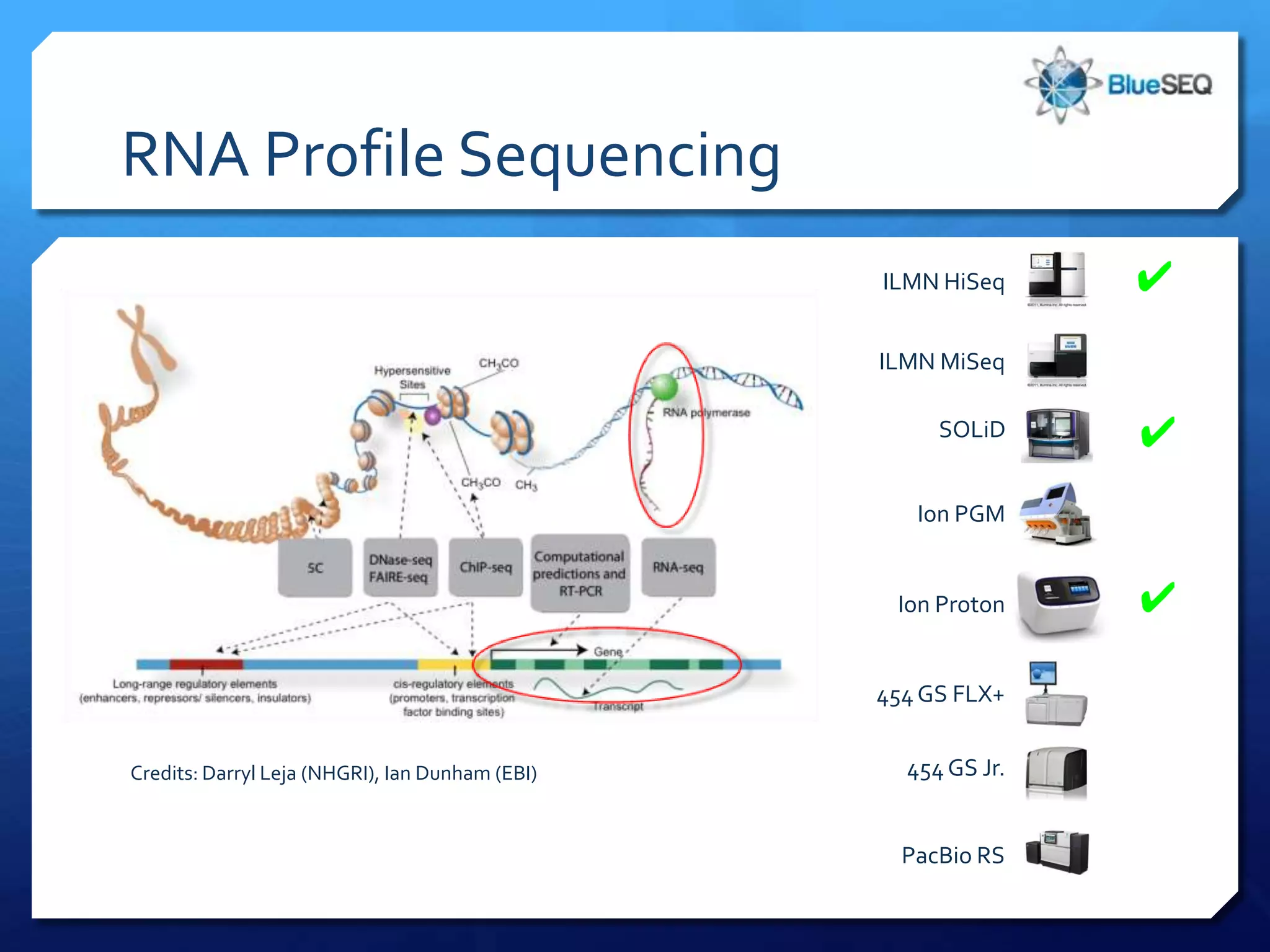
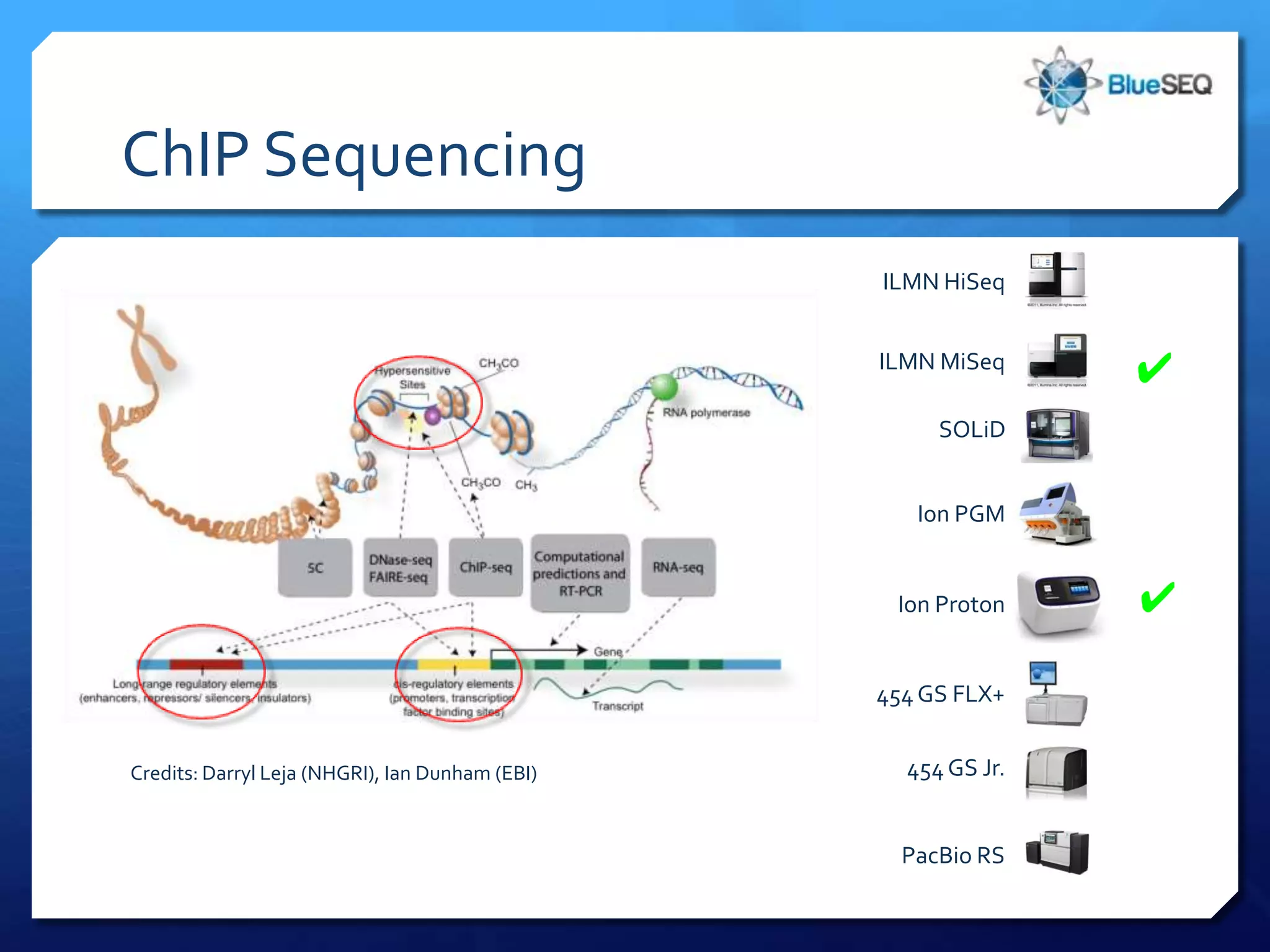
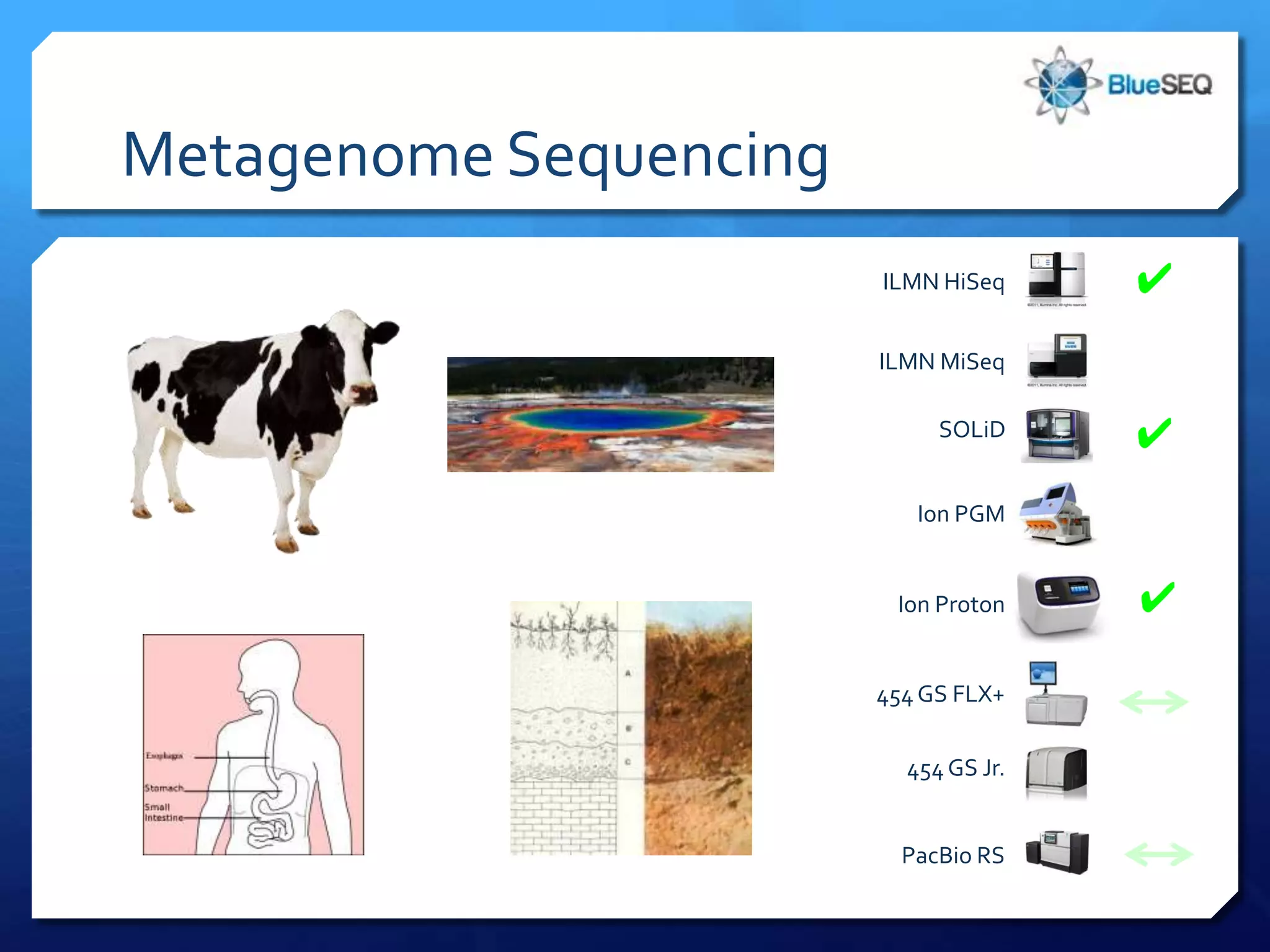
The document provides an overview of next-generation sequencing (NGS) platforms, detailing applications, performance specifications, and future trends. Key highlights include various sequencing types like whole genome, exome, small genome, and targeted sequencing, along with comparisons of different platform technologies such as Illumina, Life Technologies, and Ion Torrent. Major trends indicate a shift towards desktop machines, faster runs, and increased usage in diagnostic applications.