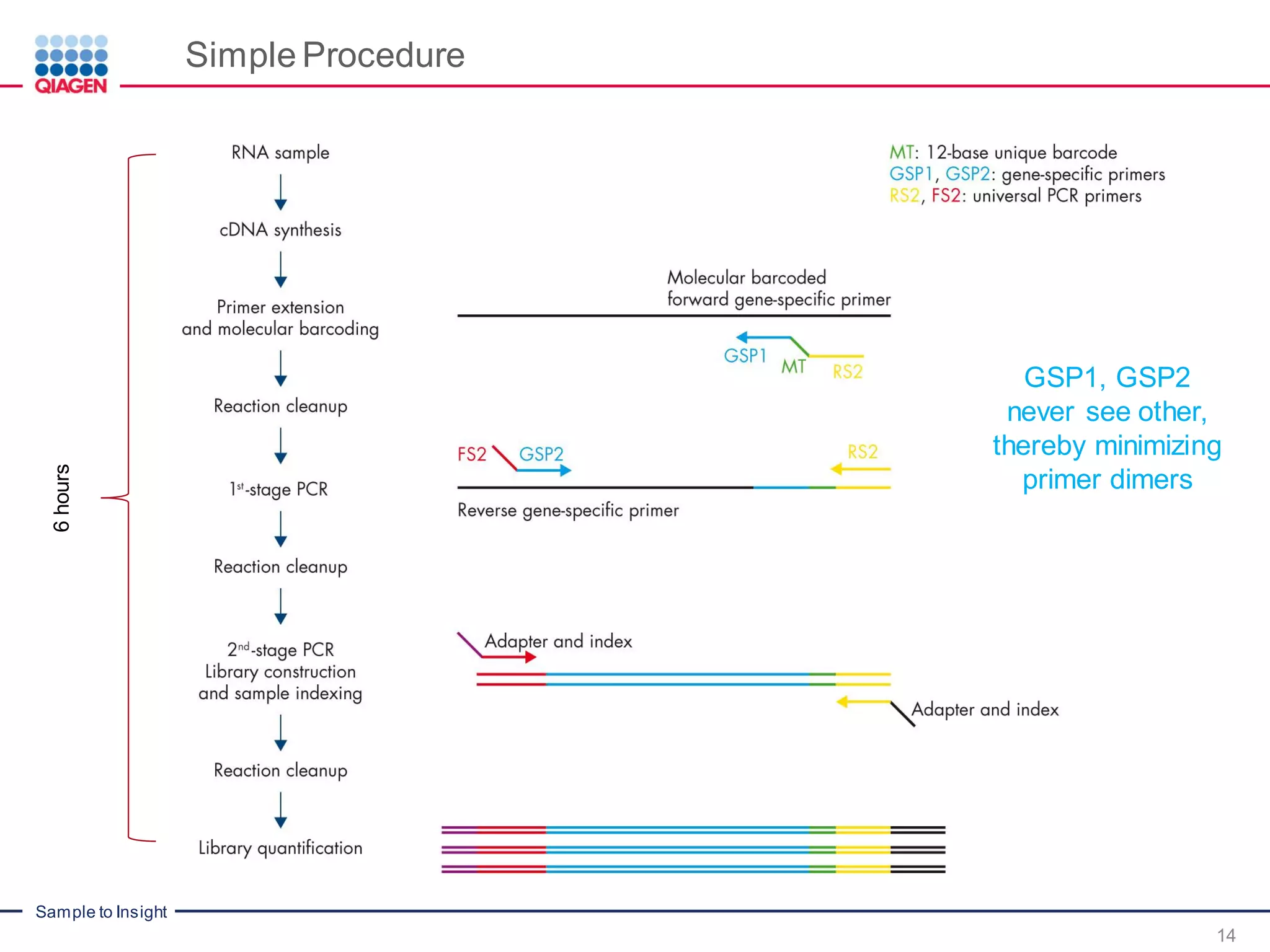
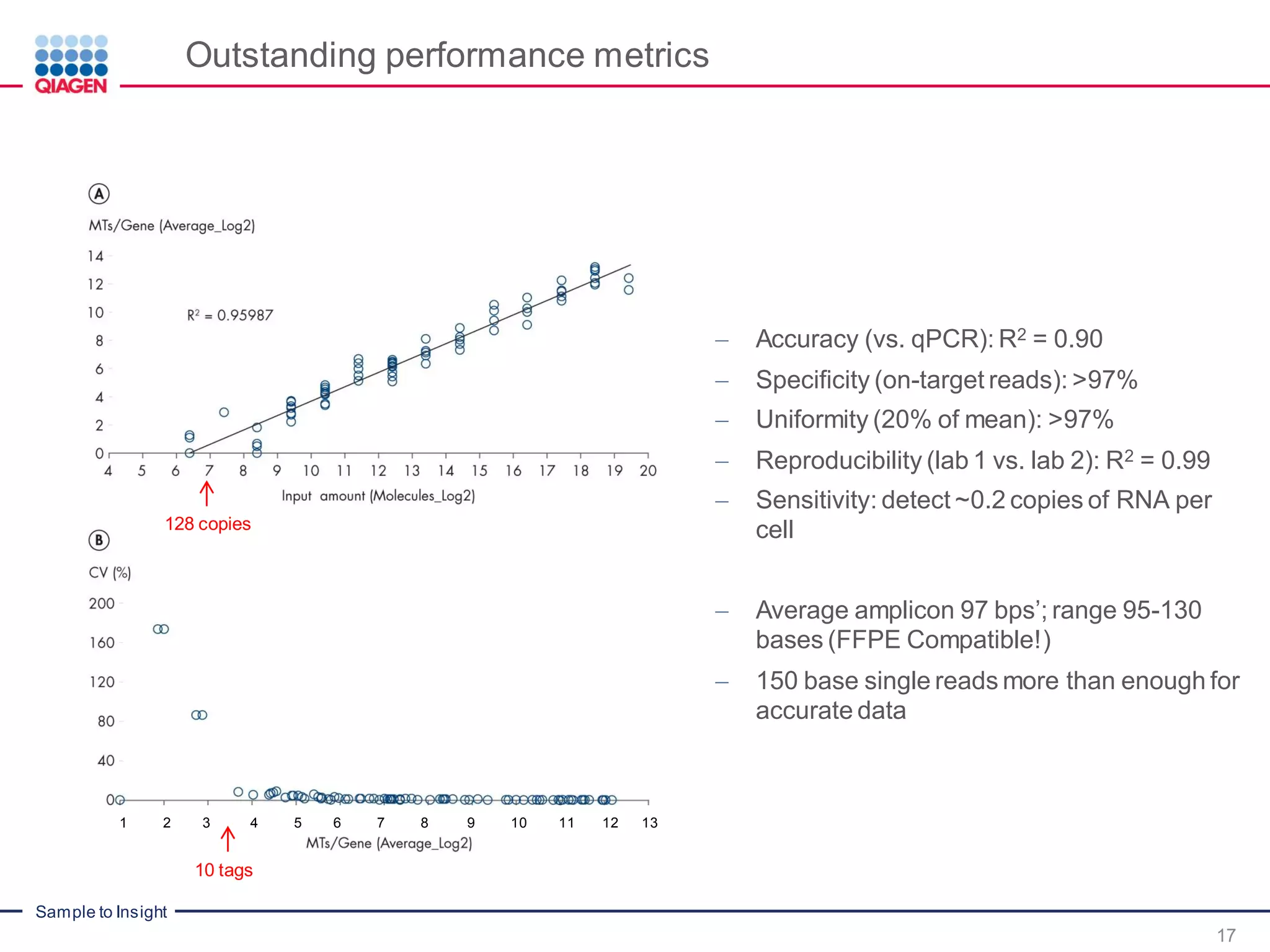
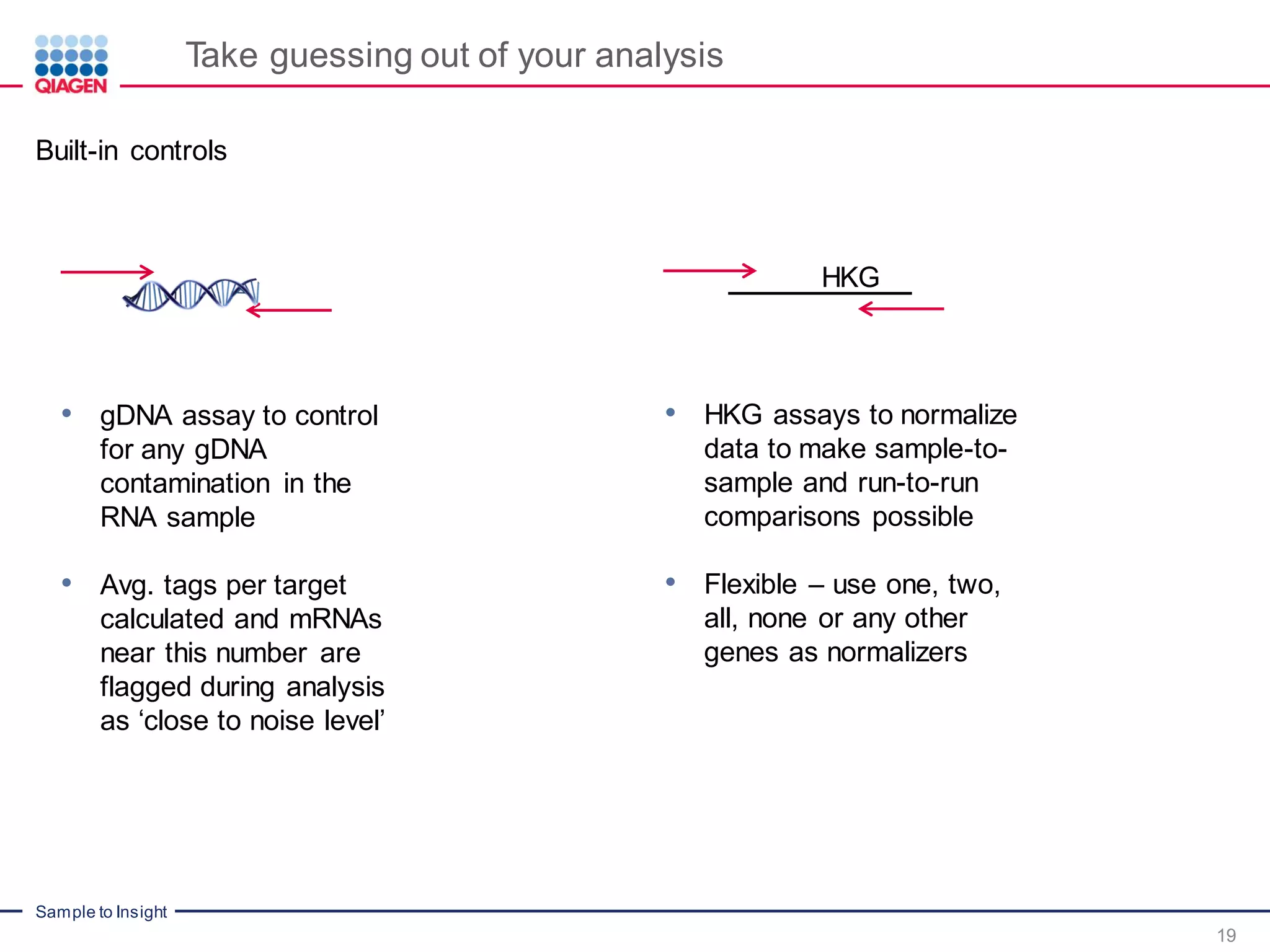
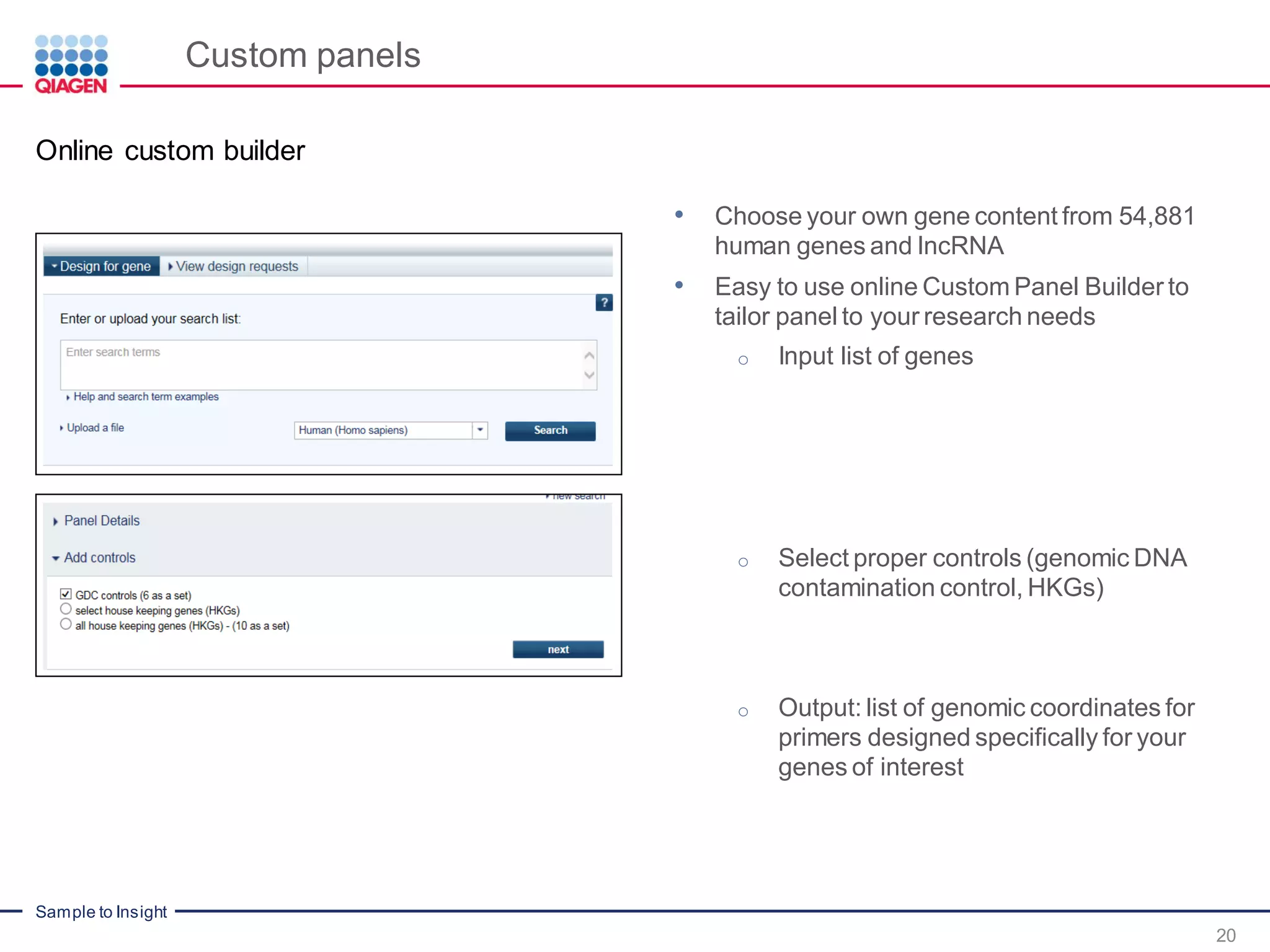
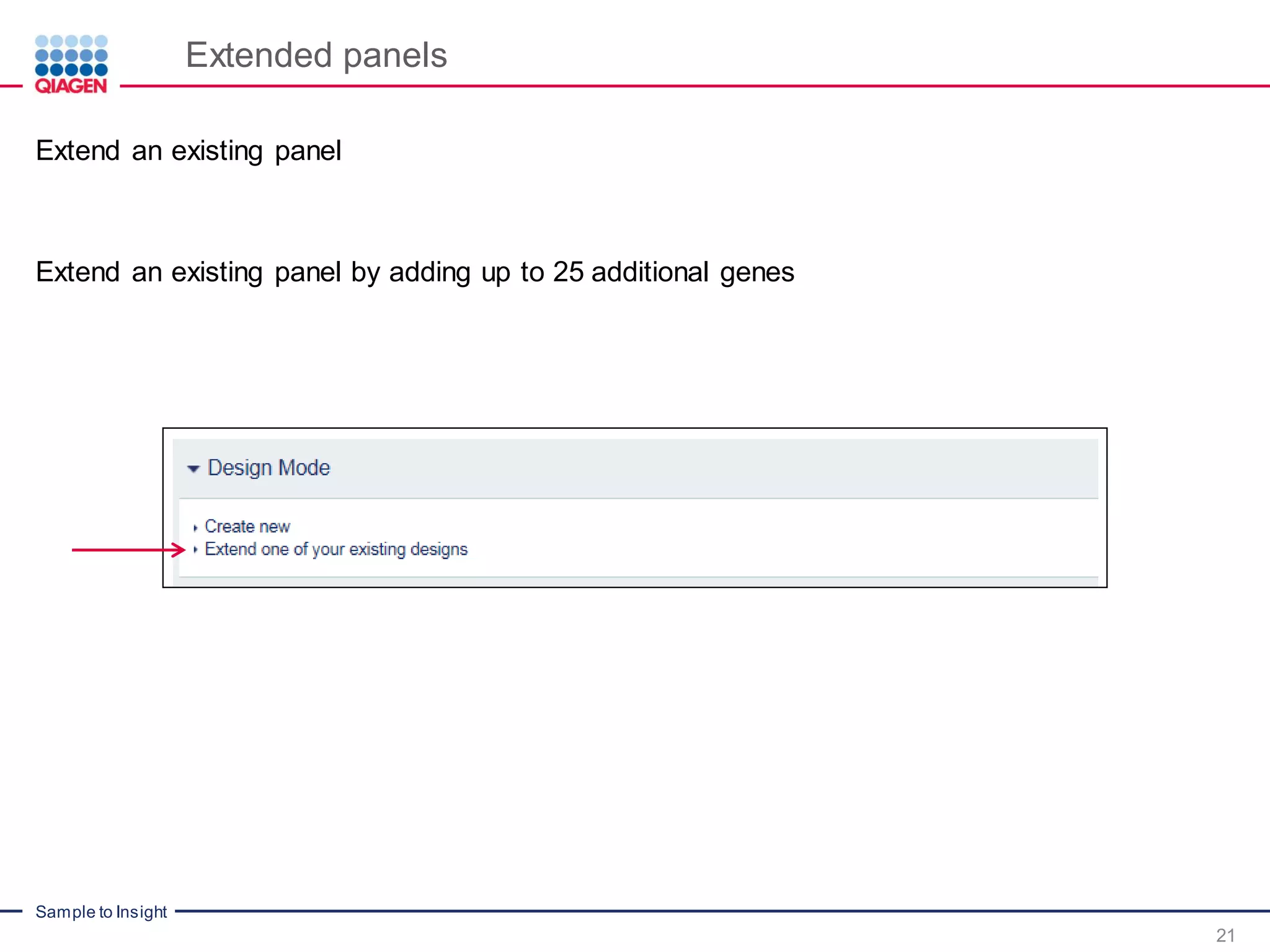
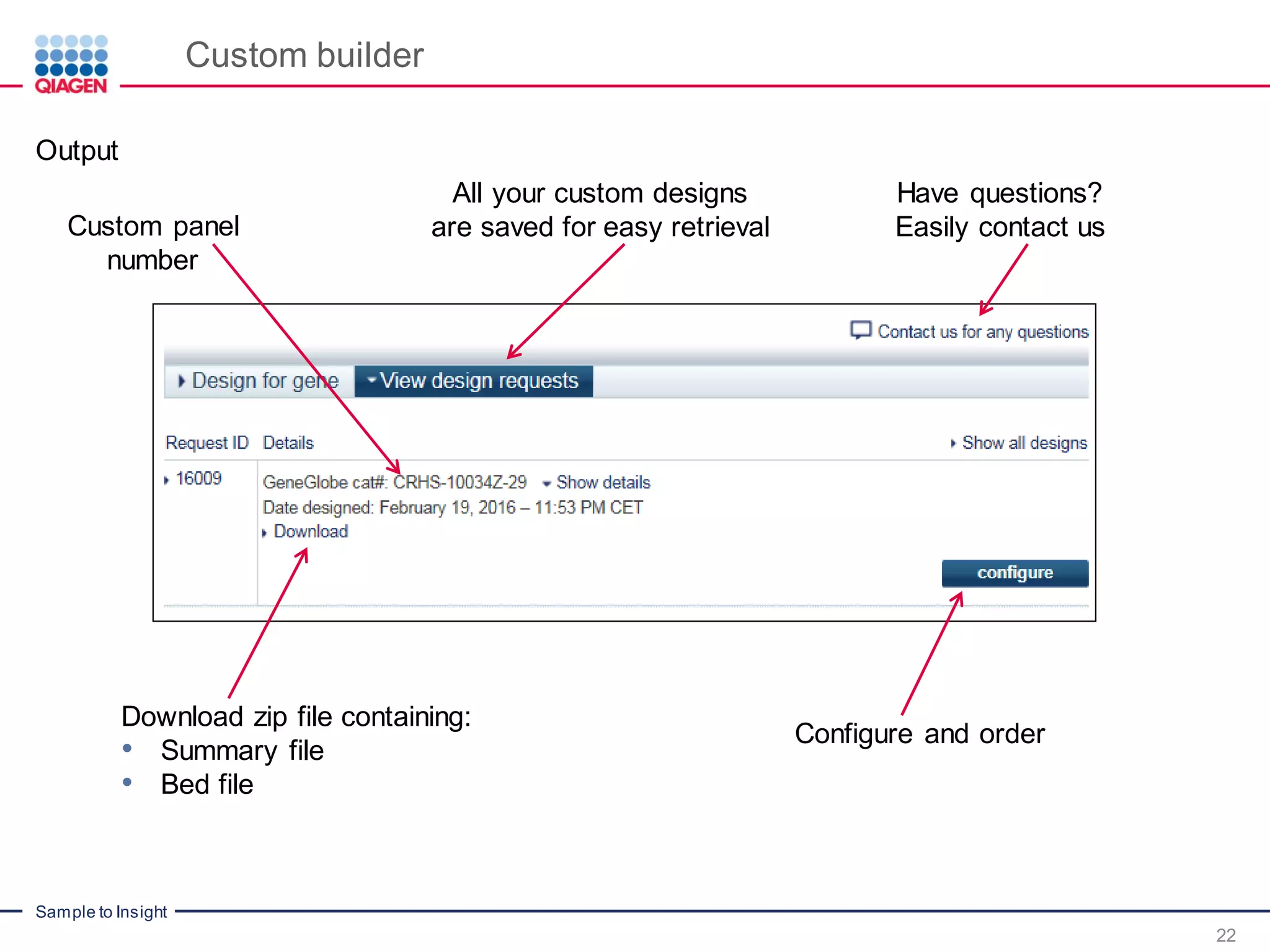
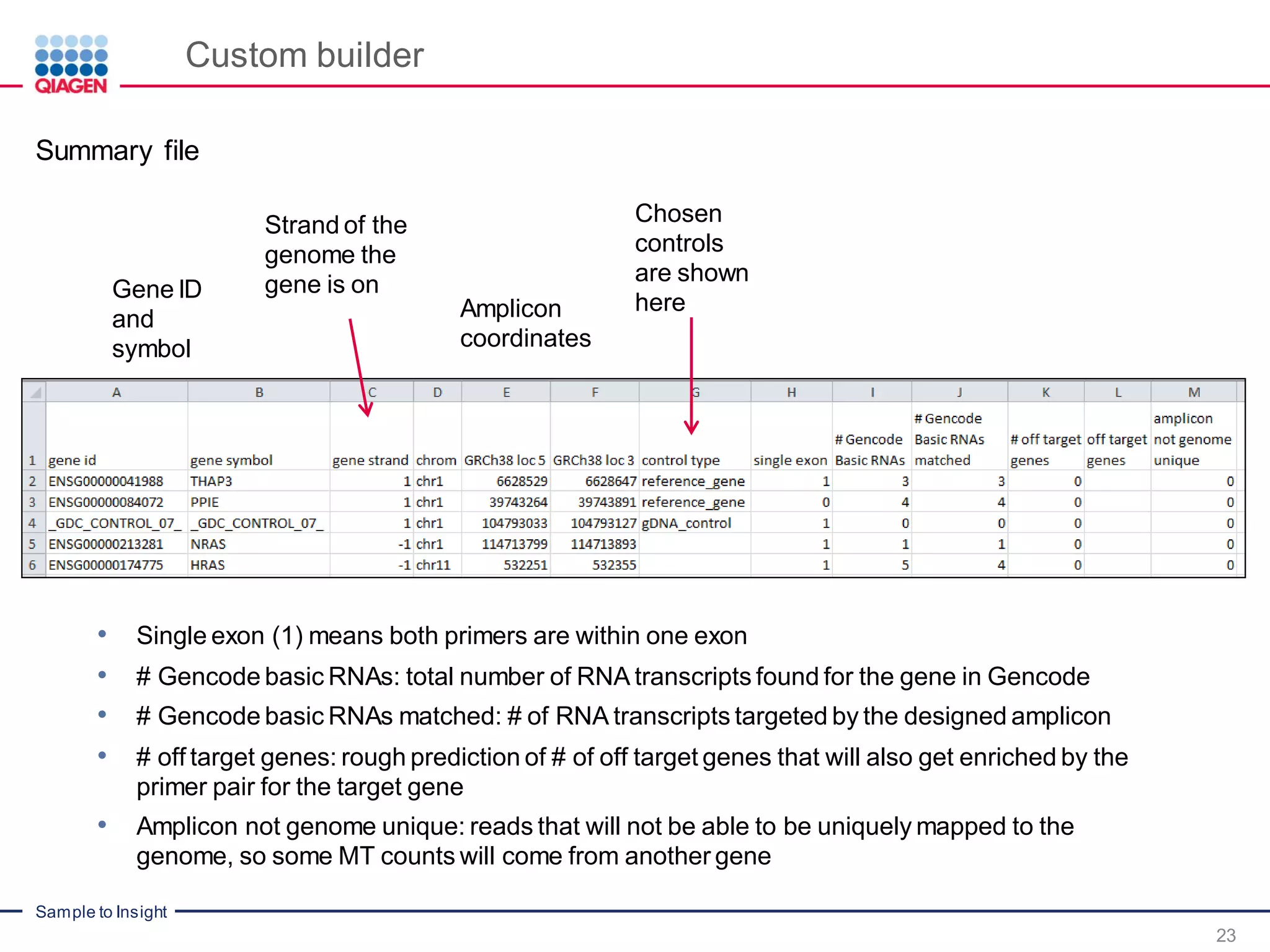
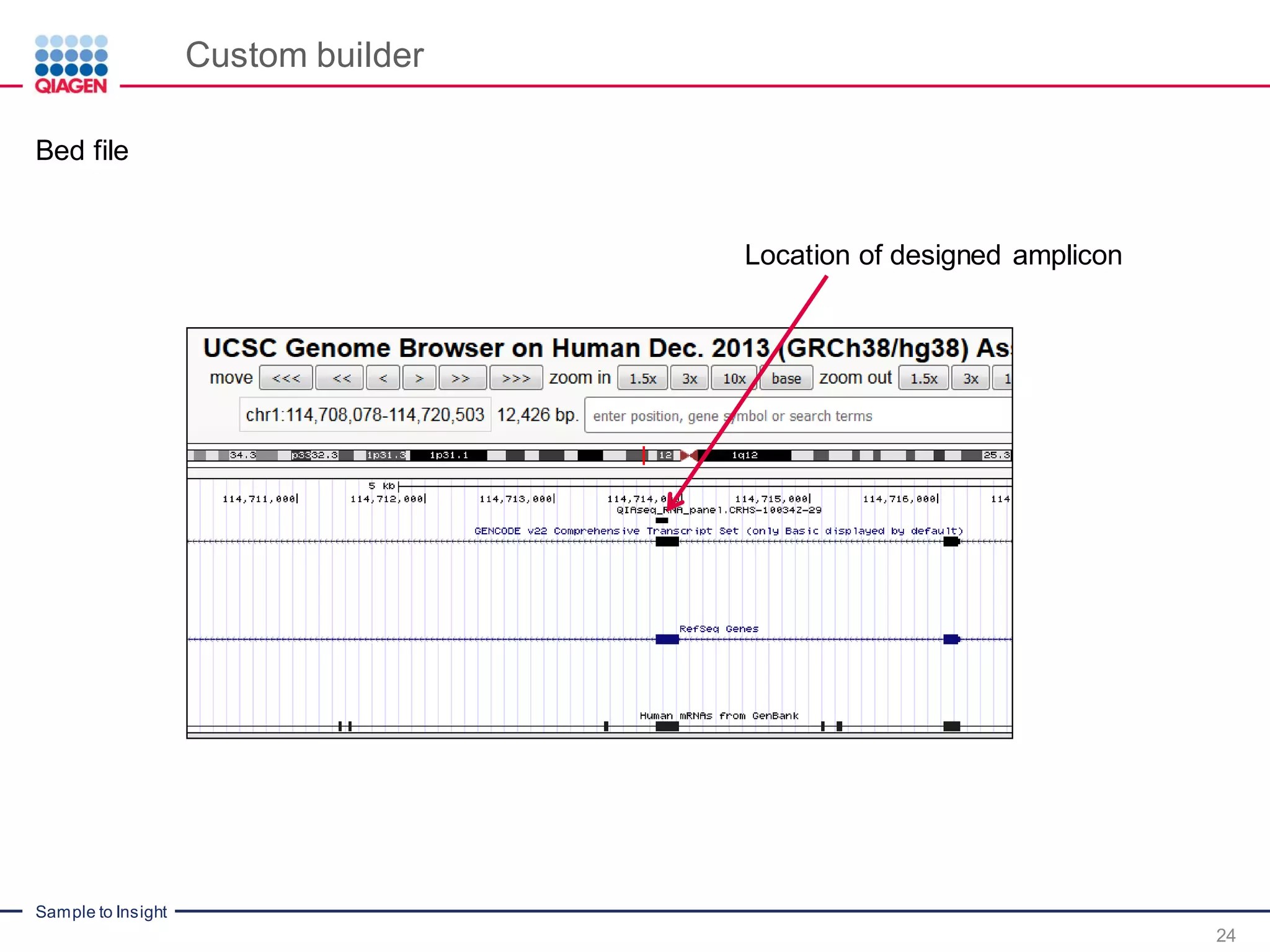
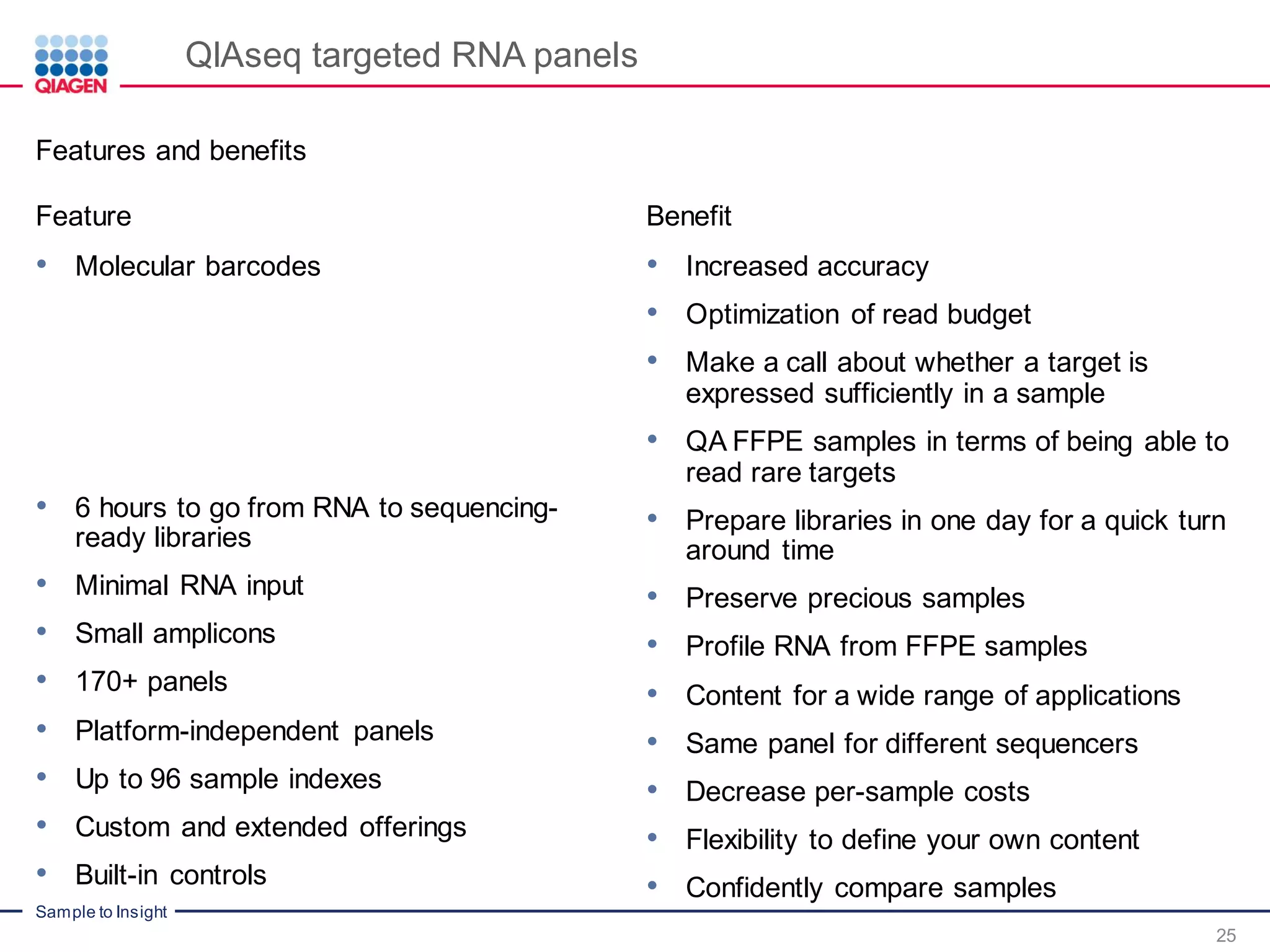
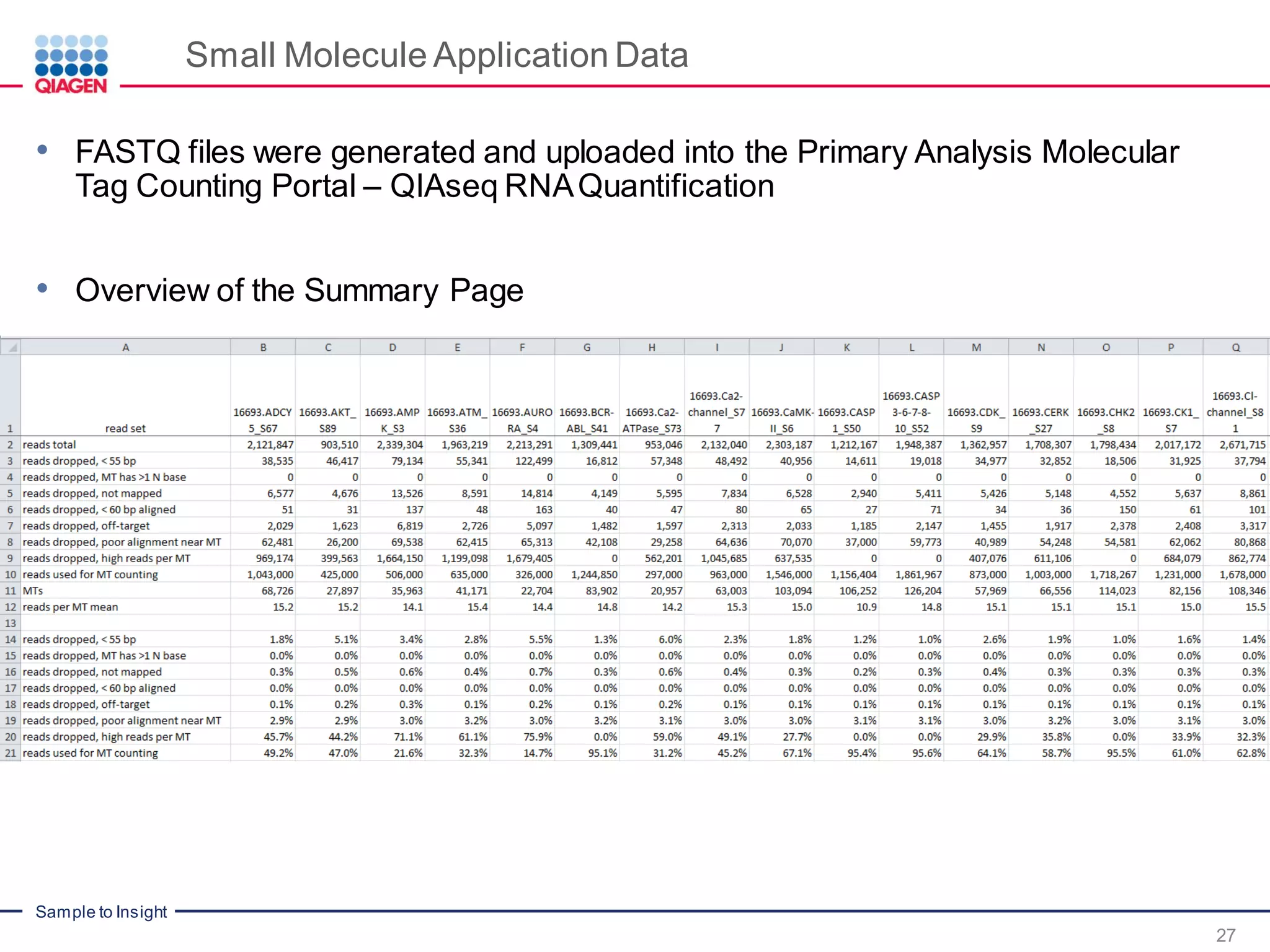
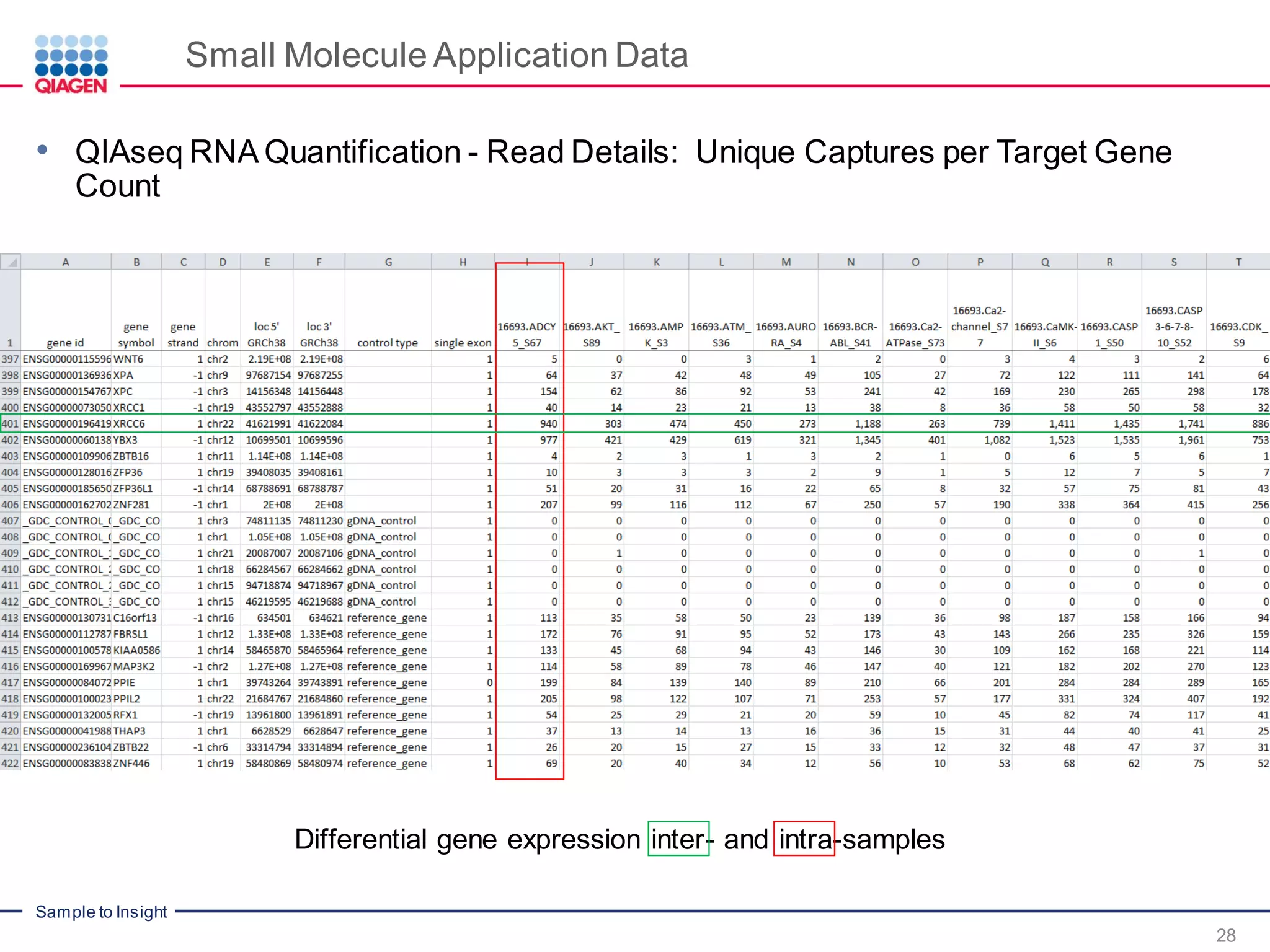
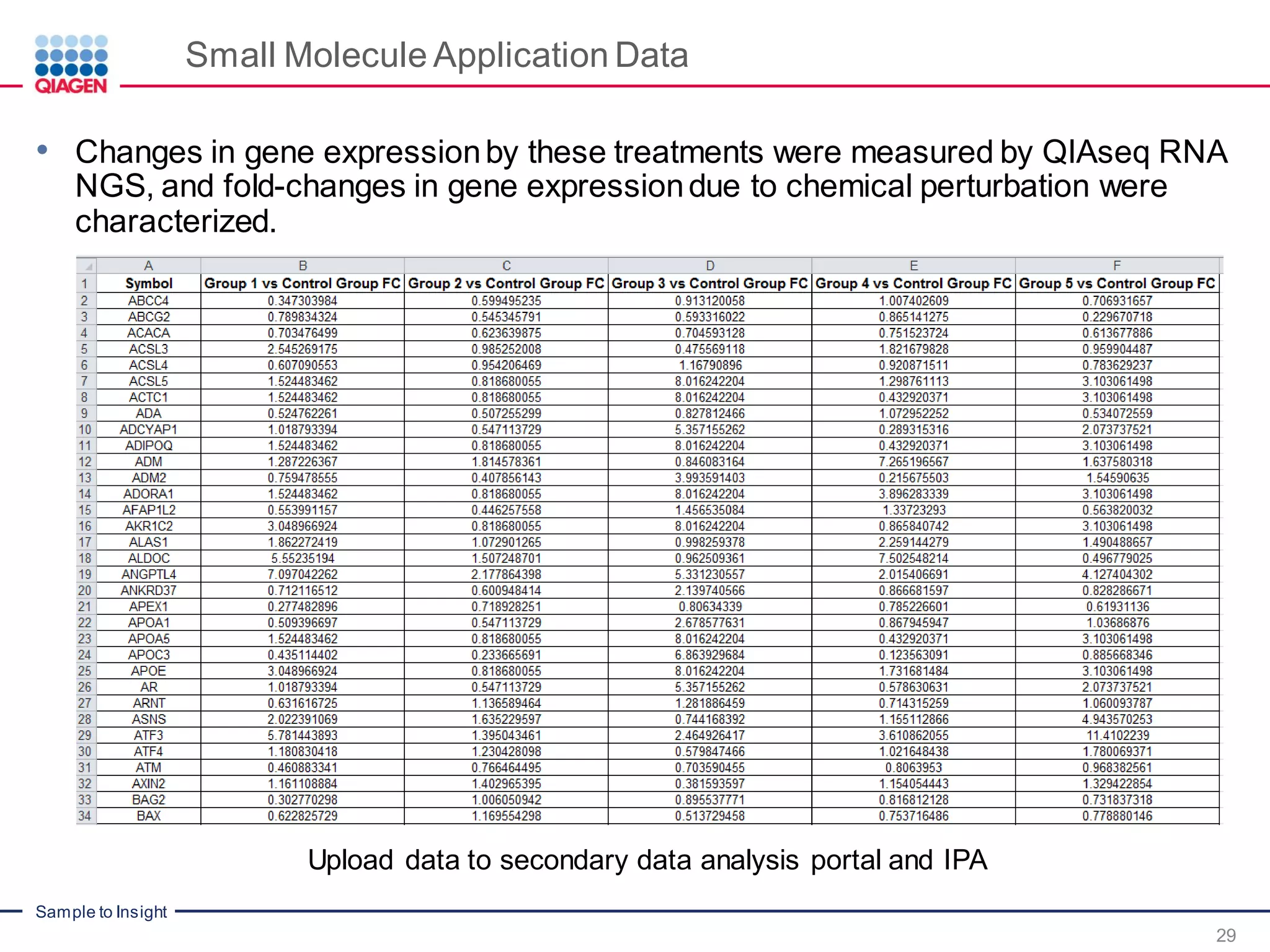
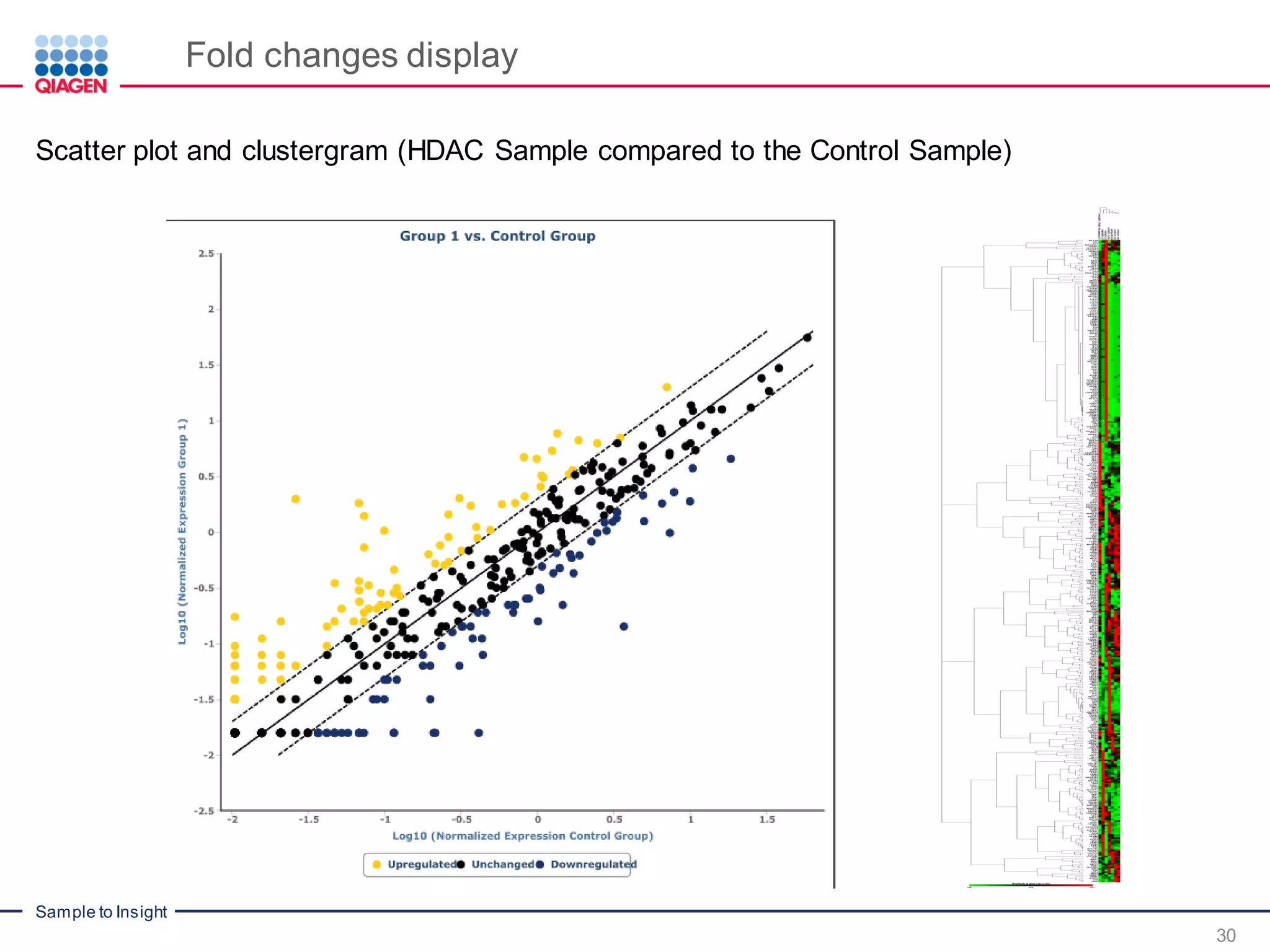
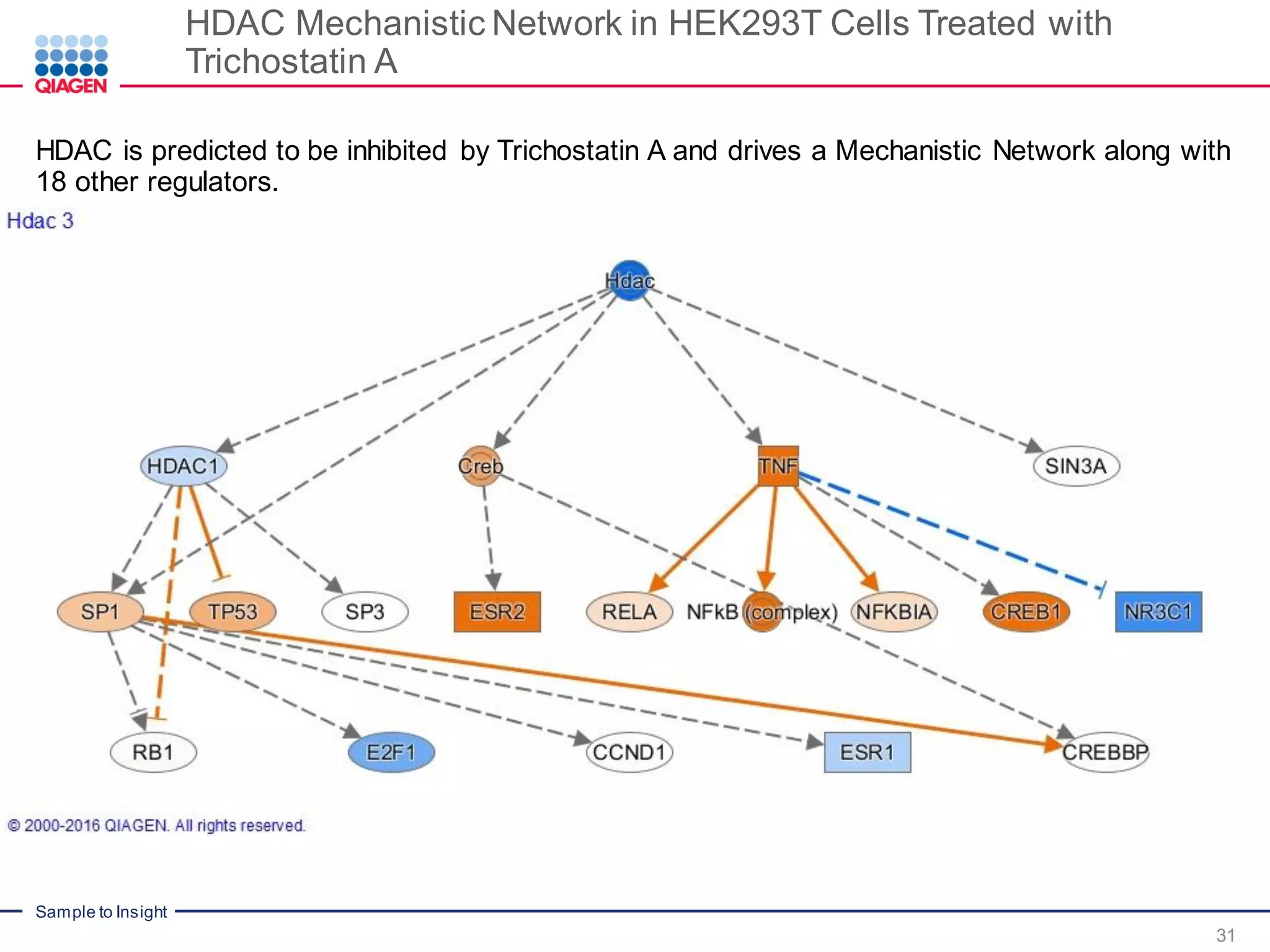
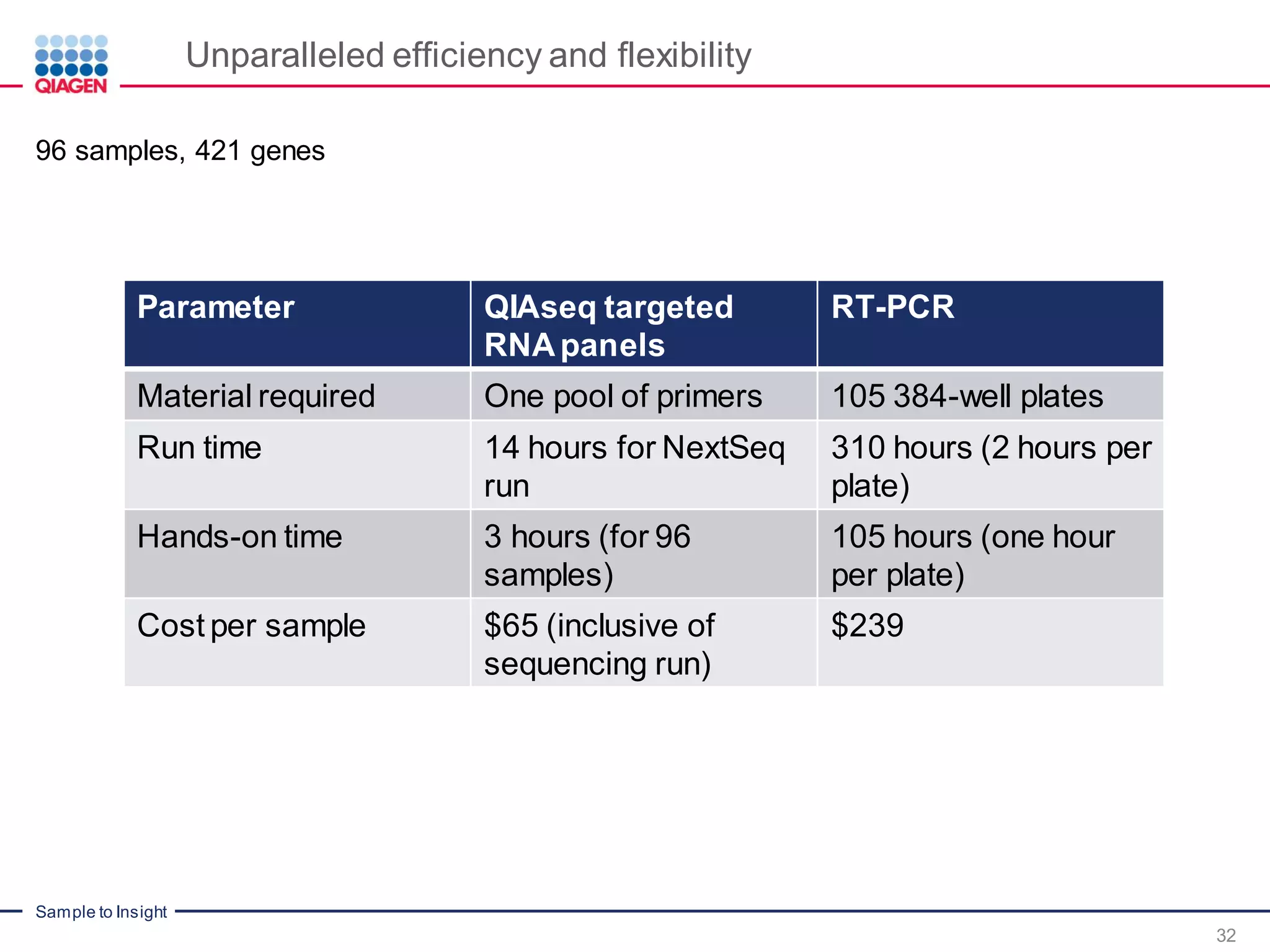
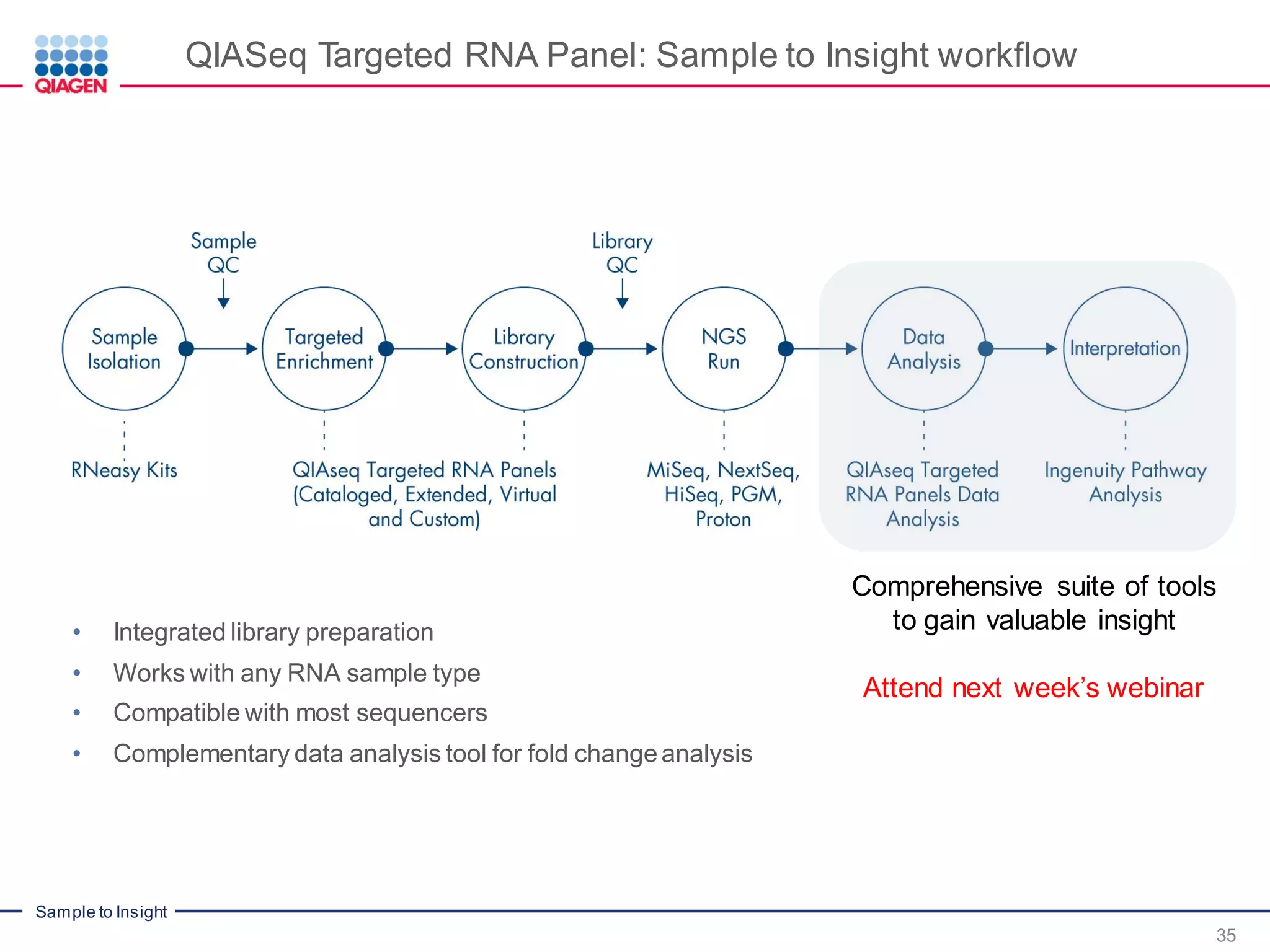
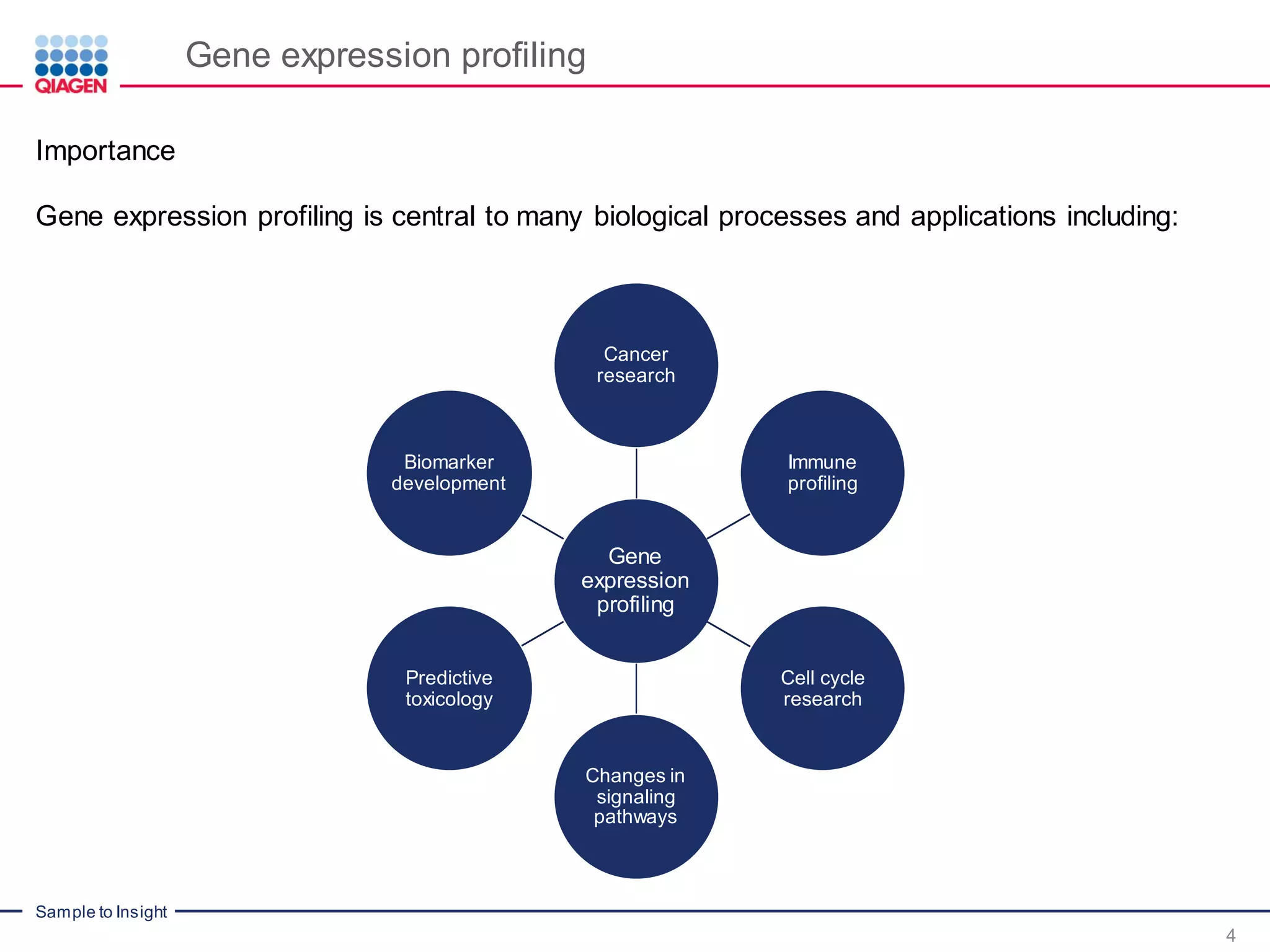
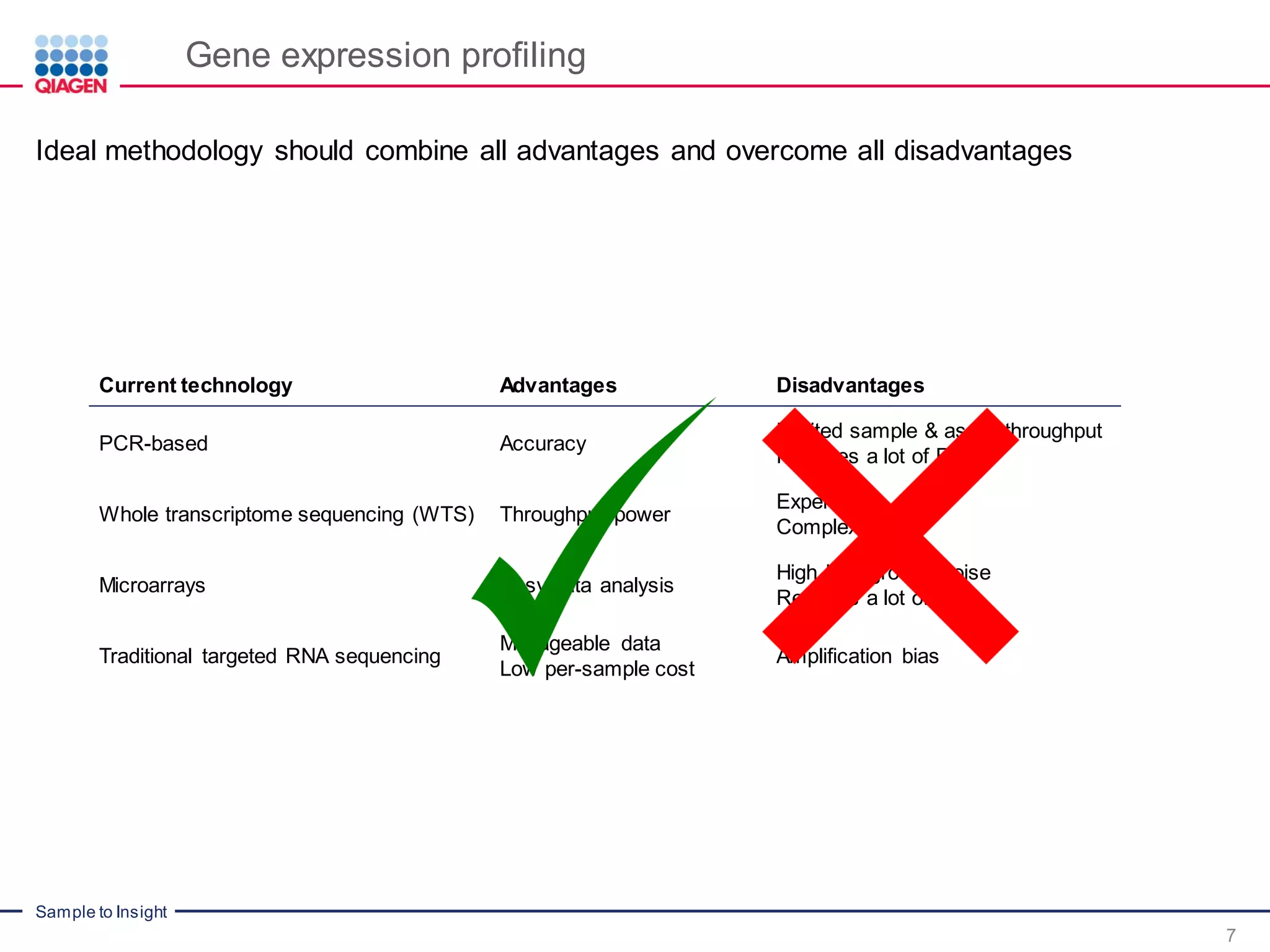
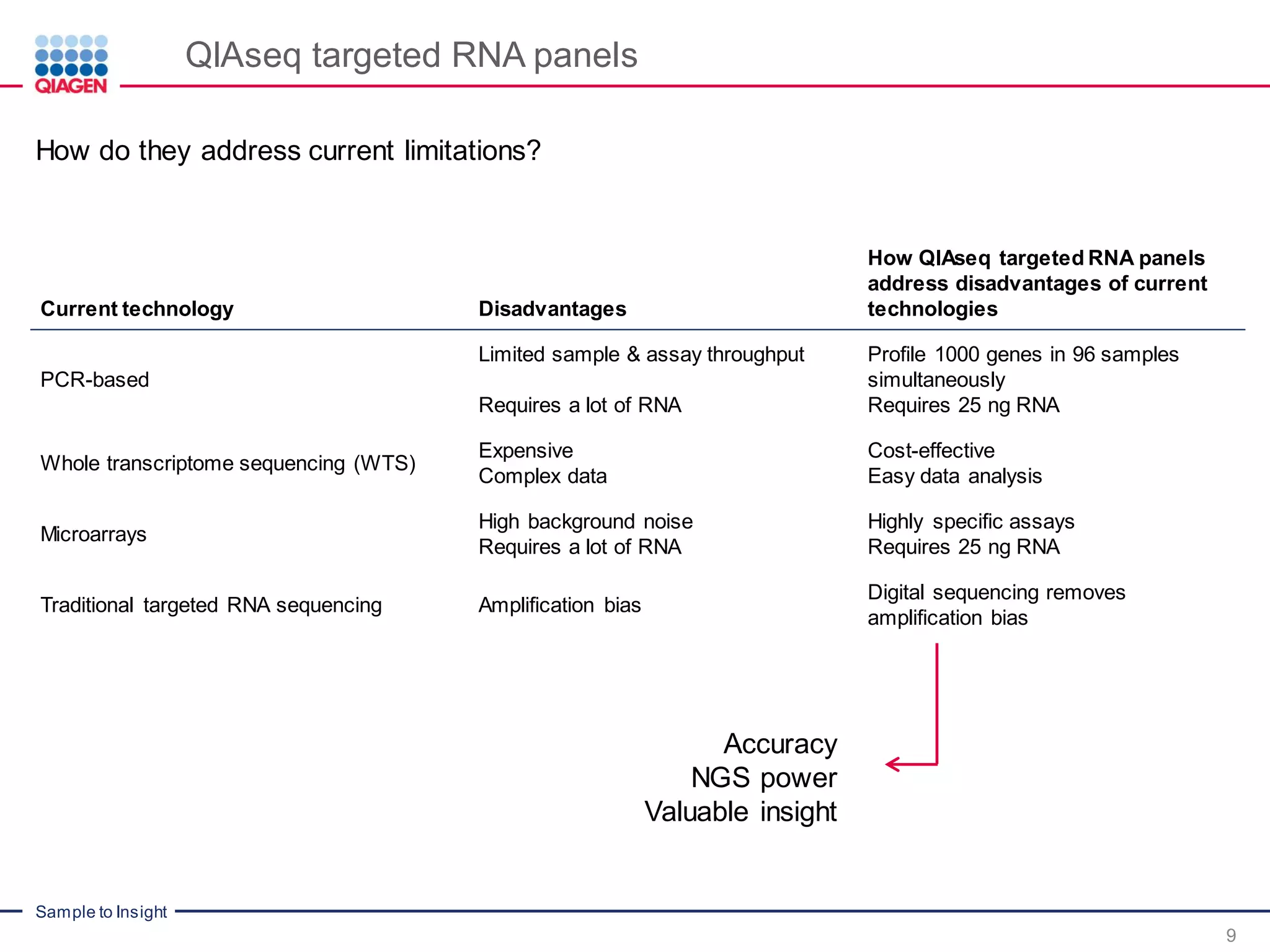
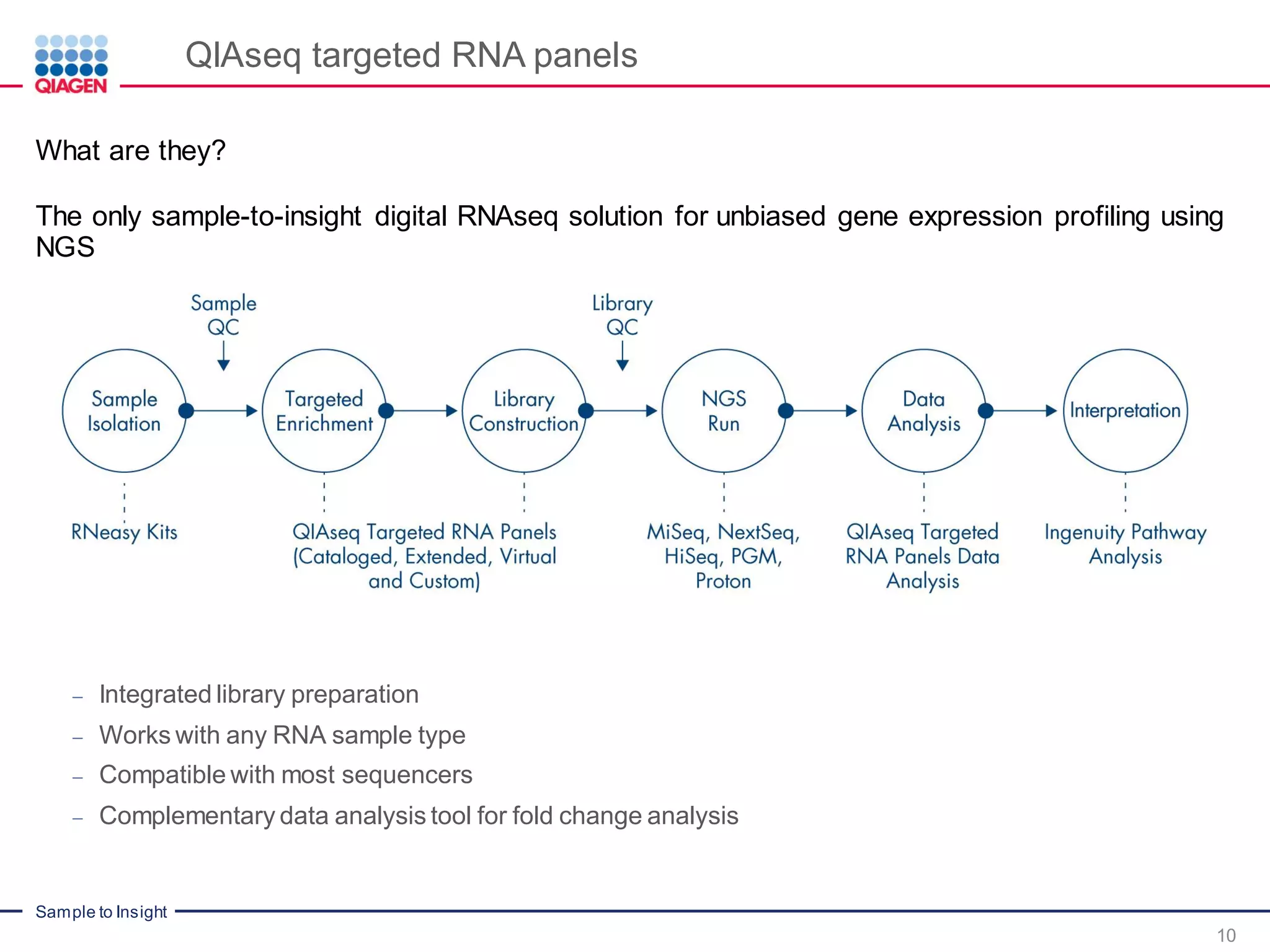
The document outlines a three-part webinar series on digital RNA sequencing (RNA-seq) aimed at accurate gene expression profiling, featuring speakers from Qiagen. It discusses the advantages and challenges of current gene expression profiling methodologies and introduces Qiagen's targeted RNA panels as a comprehensive solution to improve accuracy while overcoming limitations of traditional methods. The webinars include tutorials on data analysis and applications in various research fields, emphasizing the effectiveness of molecular barcoding technology in achieving unbiased gene expression results.


![Sample to Insight
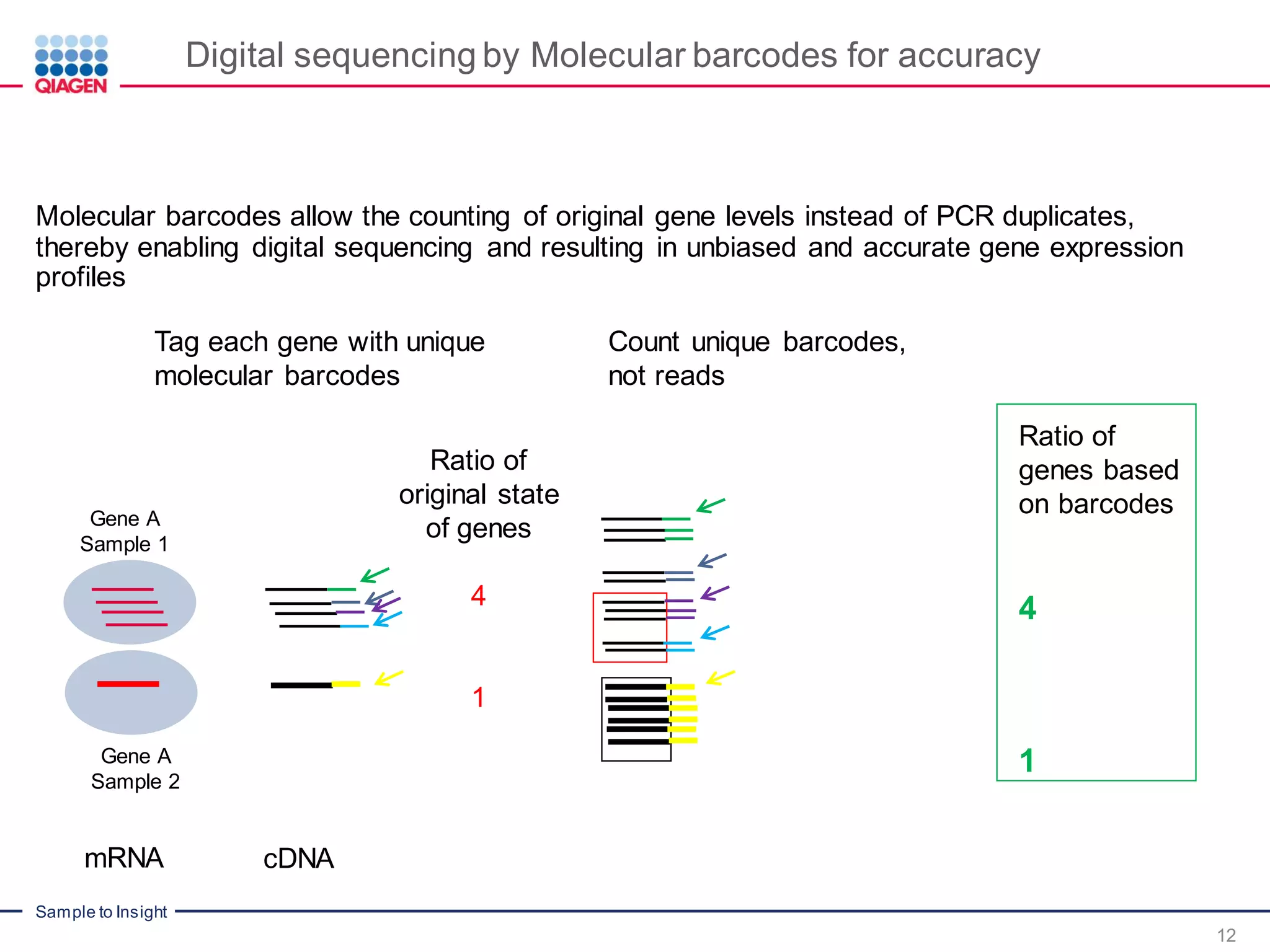
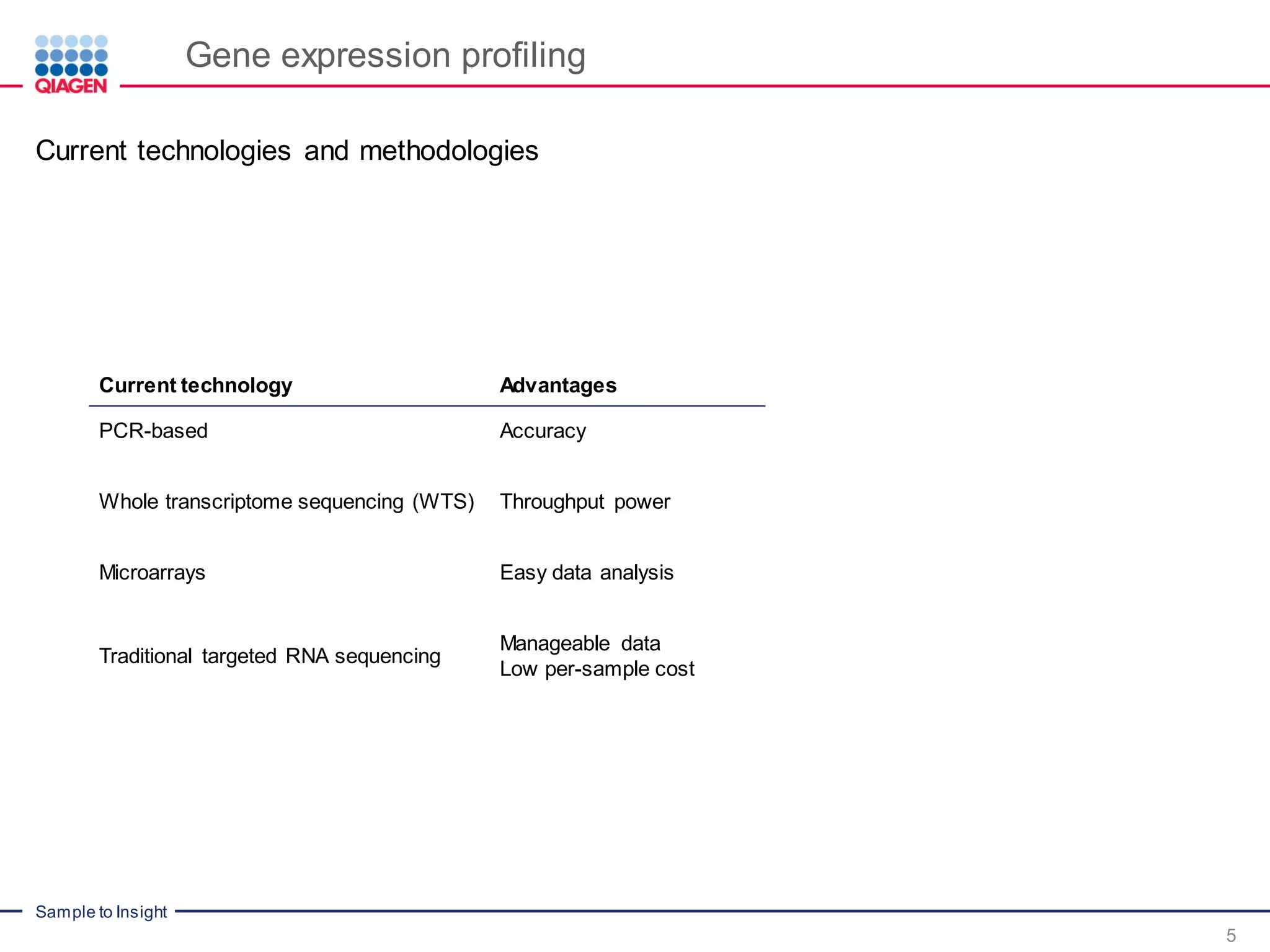
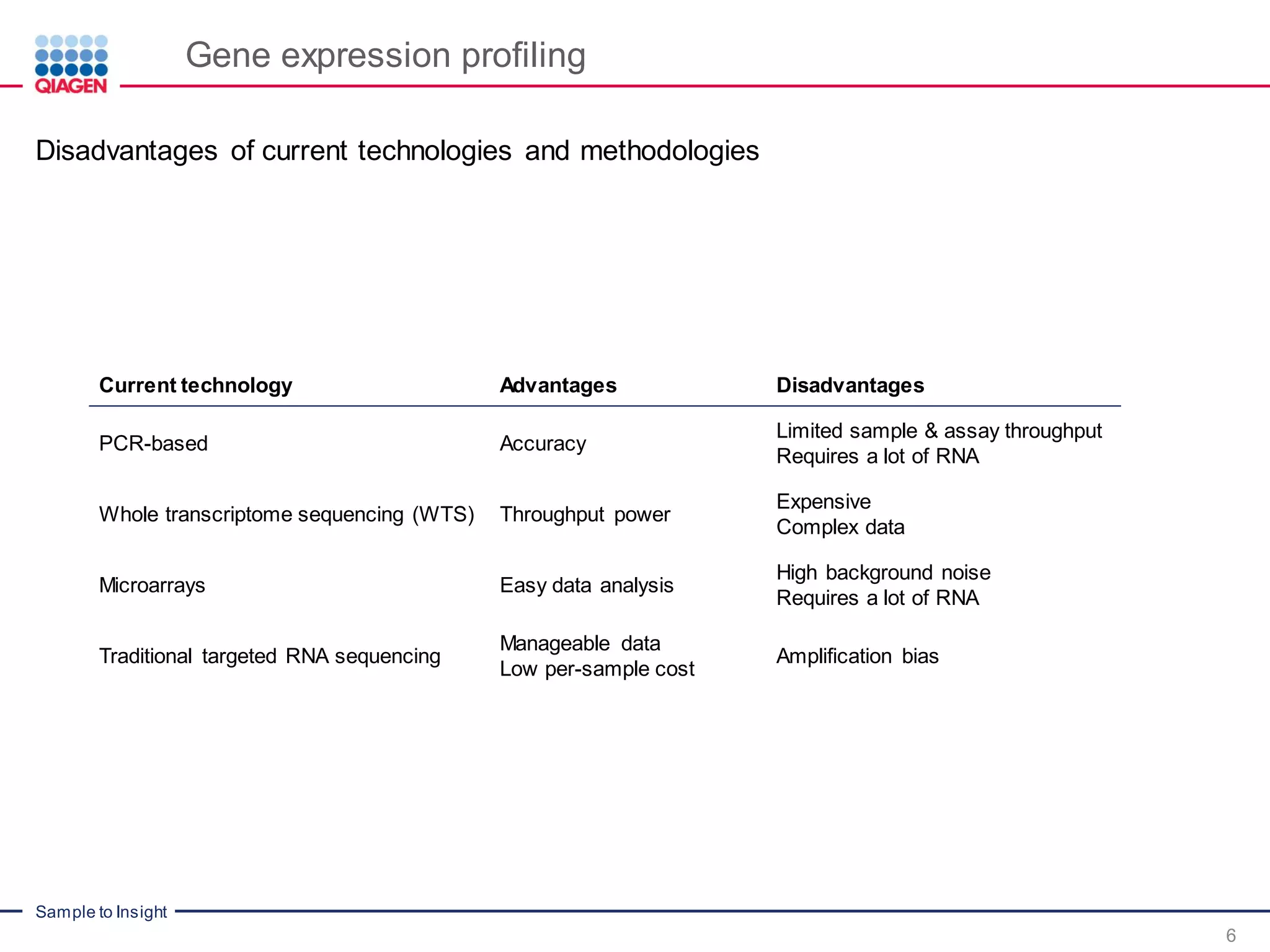
Digital sequencing by Molecular barcodes for accuracy
11
PCR duplicates and amplification bias are major issues in current RNAseq workflows, as they
result in biased and inaccurate gene expression profiles
mRNA cDNA
PCR duplicates
Ratio of
original state
of genes
4
1
Amplification bias
Ratio of genes
based
on reads
[reads (ratio)]
12 (2)
6 (1)
Ratio based on reads
Gene A
Sample 1
Gene A
Sample 2](https://image.slidesharecdn.com/rnaseqtechseriesqiaseqtargetedrnapanelsforgeneexpressionprofilingpartii-160310211447/75/Digital-RNAseq-for-Gene-Expression-Profiling-Digital-RNAseq-Webinar-Part-2-11-2048.jpg)