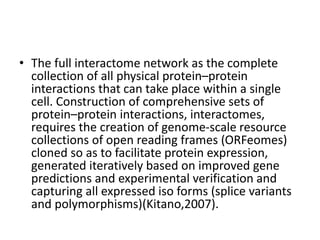
1) Interactomics is the study of interactions between genes or proteins, including genetic and physical interactions.
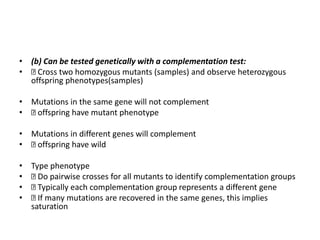
2) Complementation groups can identify mutations in the same or different genes involved in the same pathway through genetic crosses of mutant organisms.
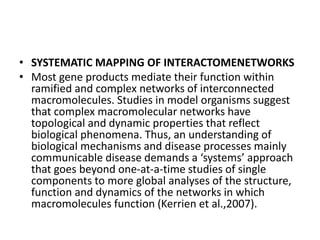
3) Modifier screens uncover new genes involved in a biological pathway by identifying mutations that alter the phenotype of an original mutant. Mapping protein-protein interaction networks provides a framework for understanding biology as an integrated system.