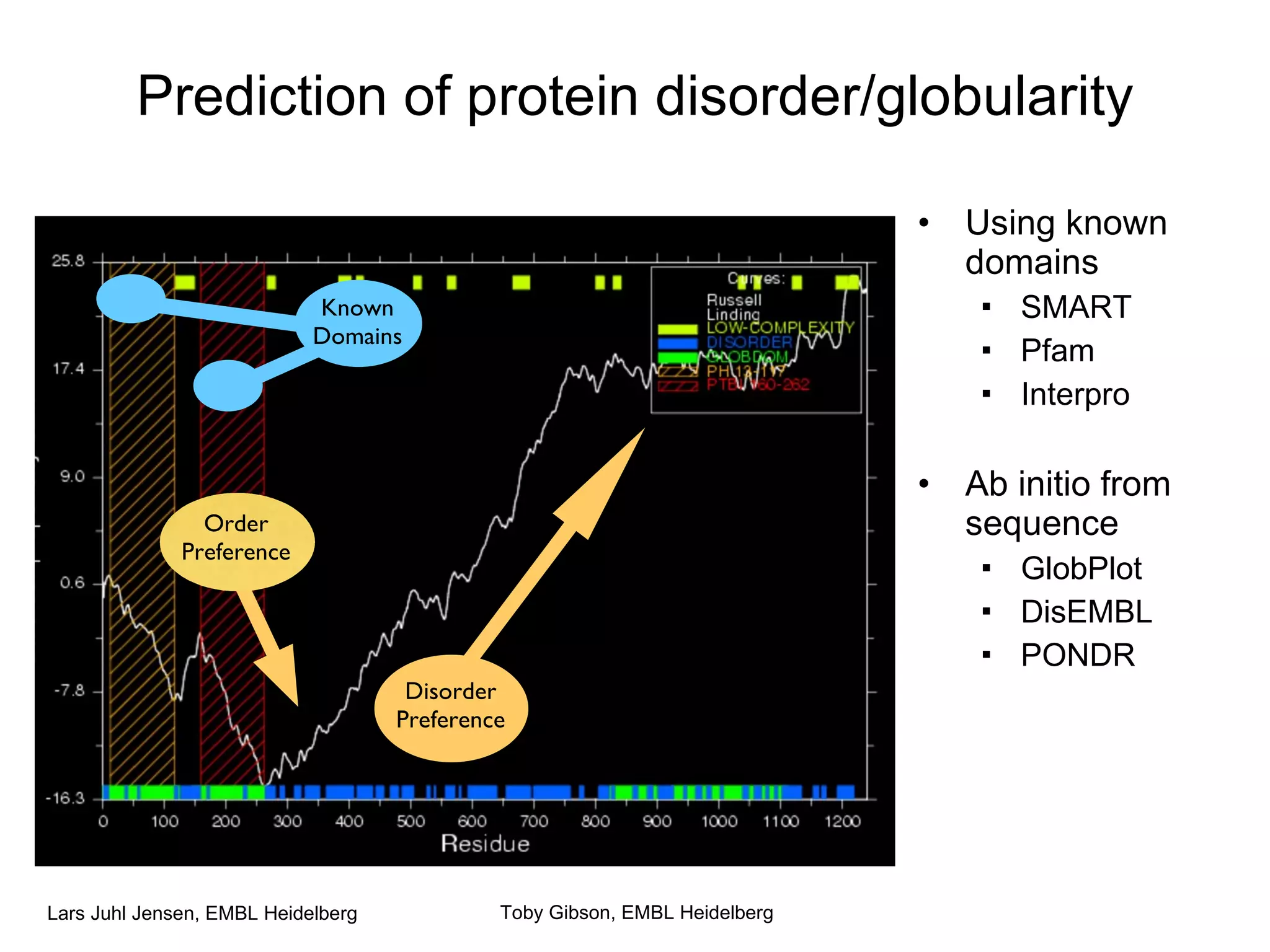
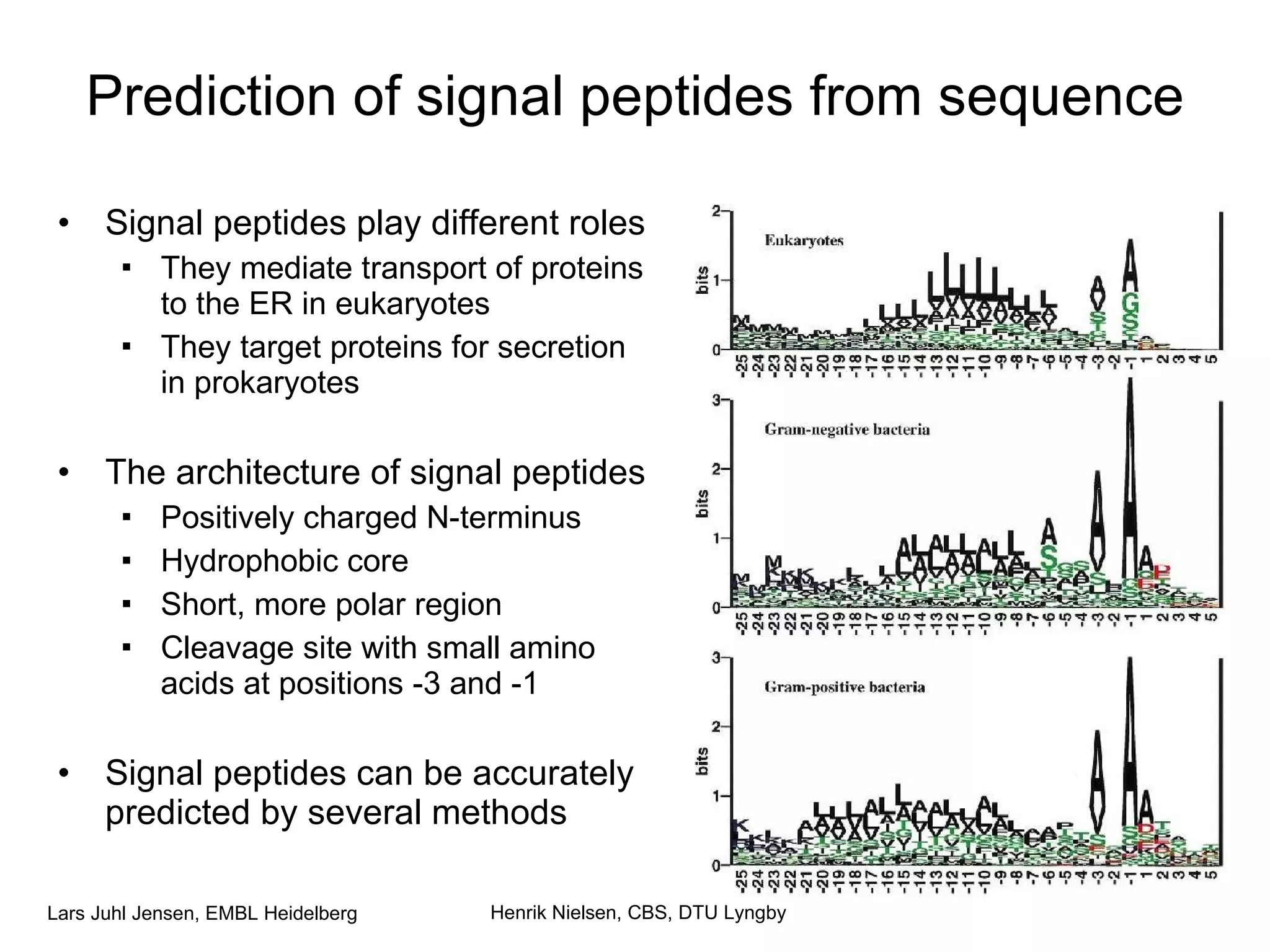
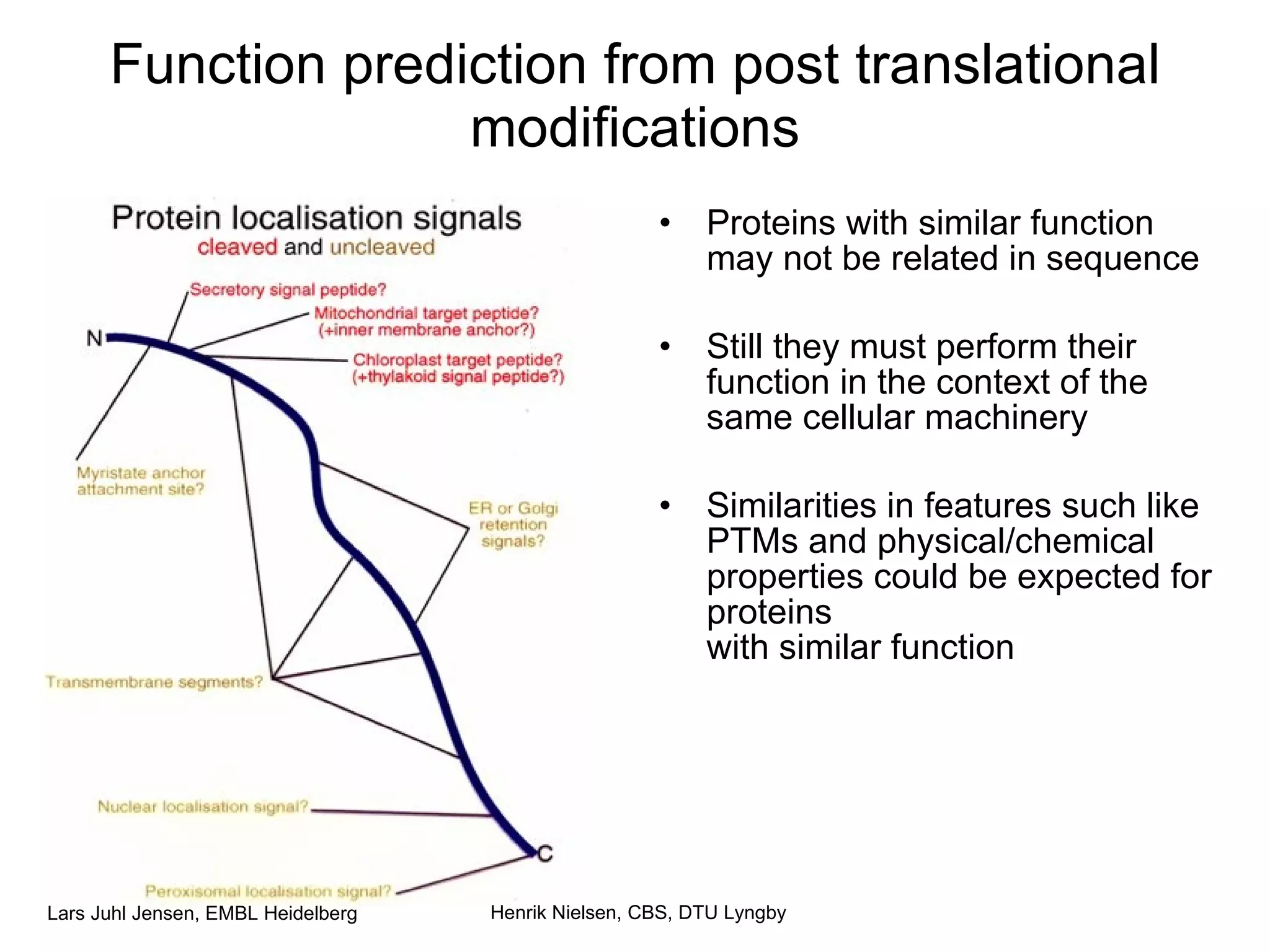
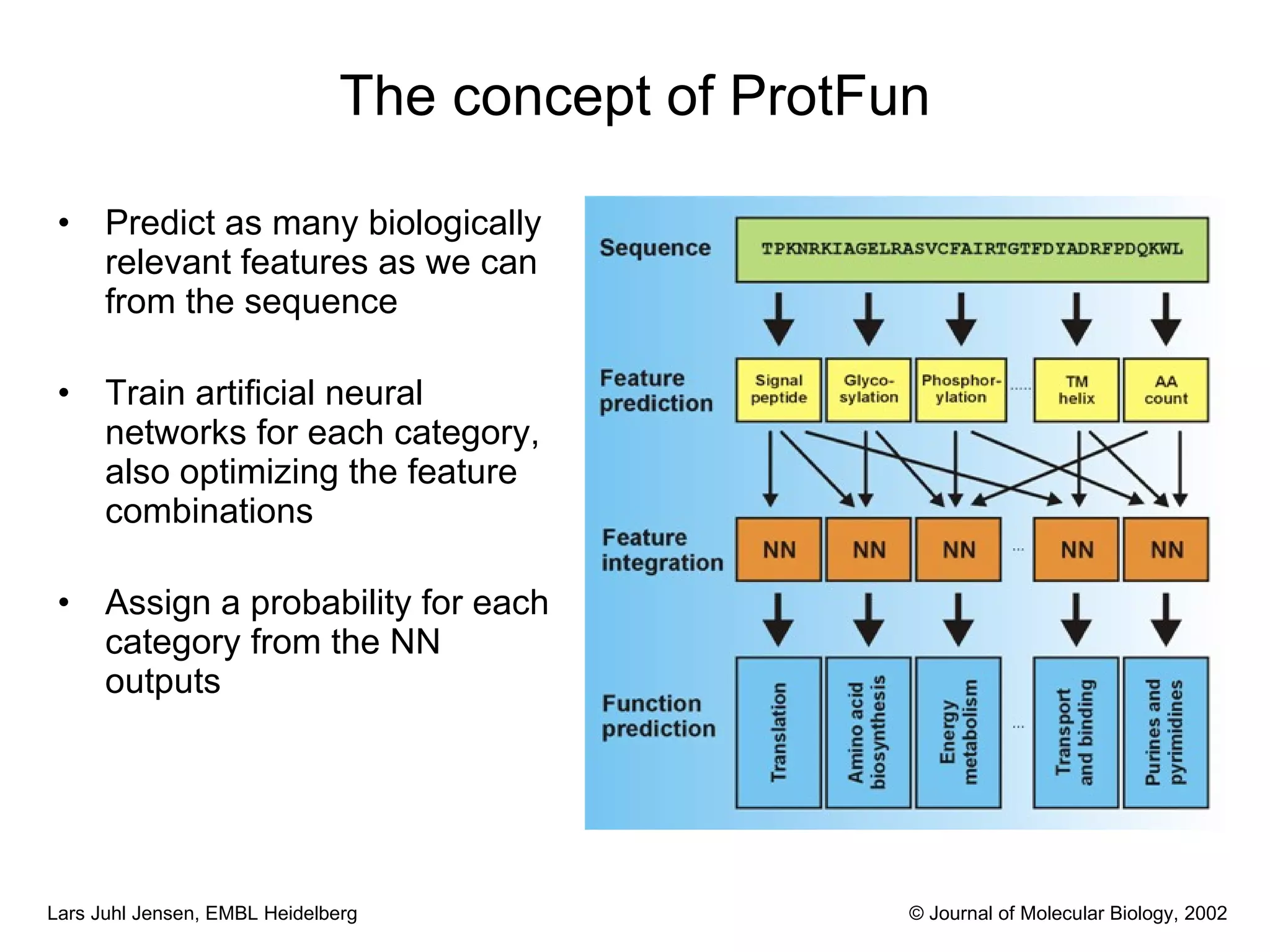
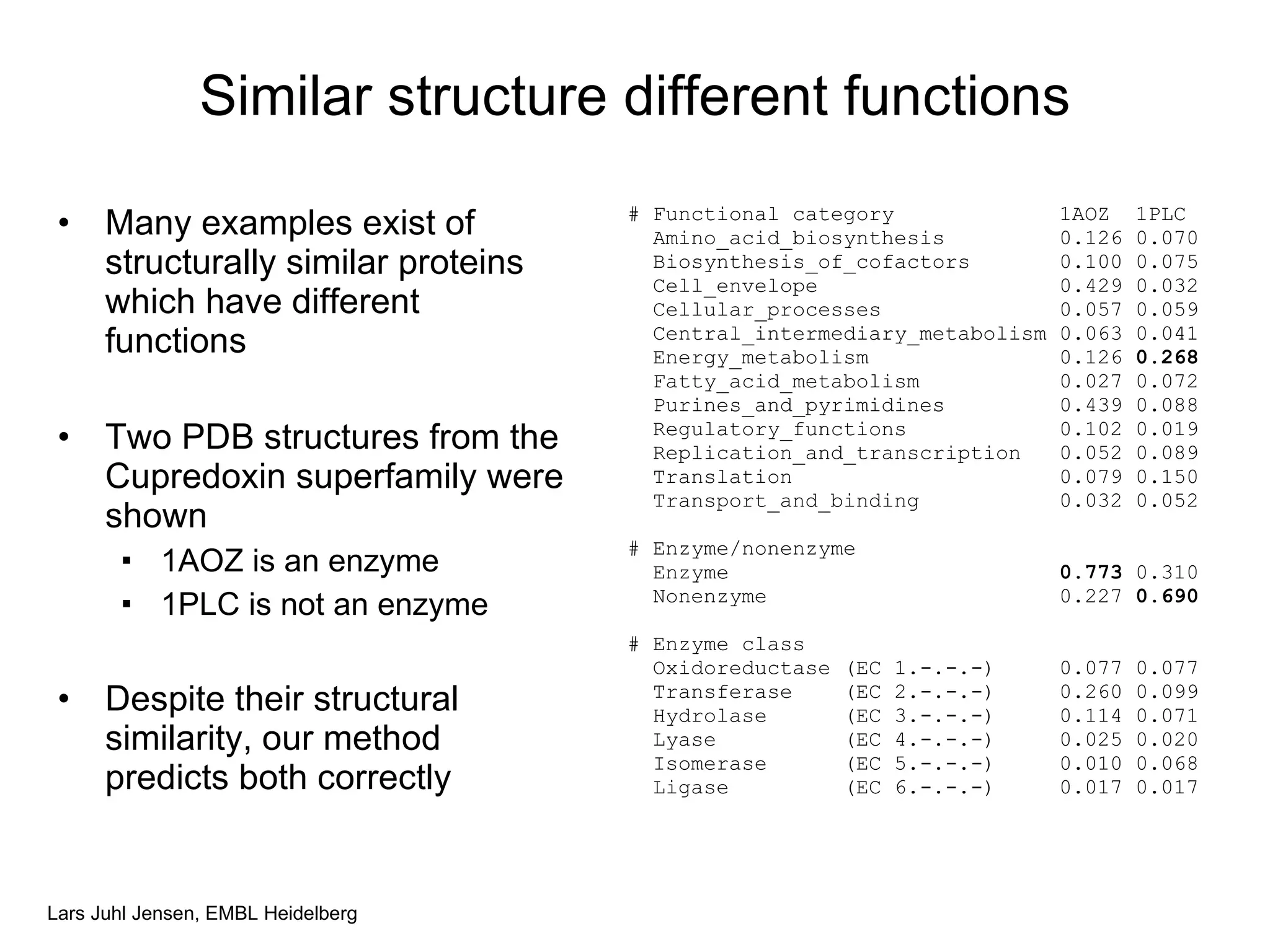
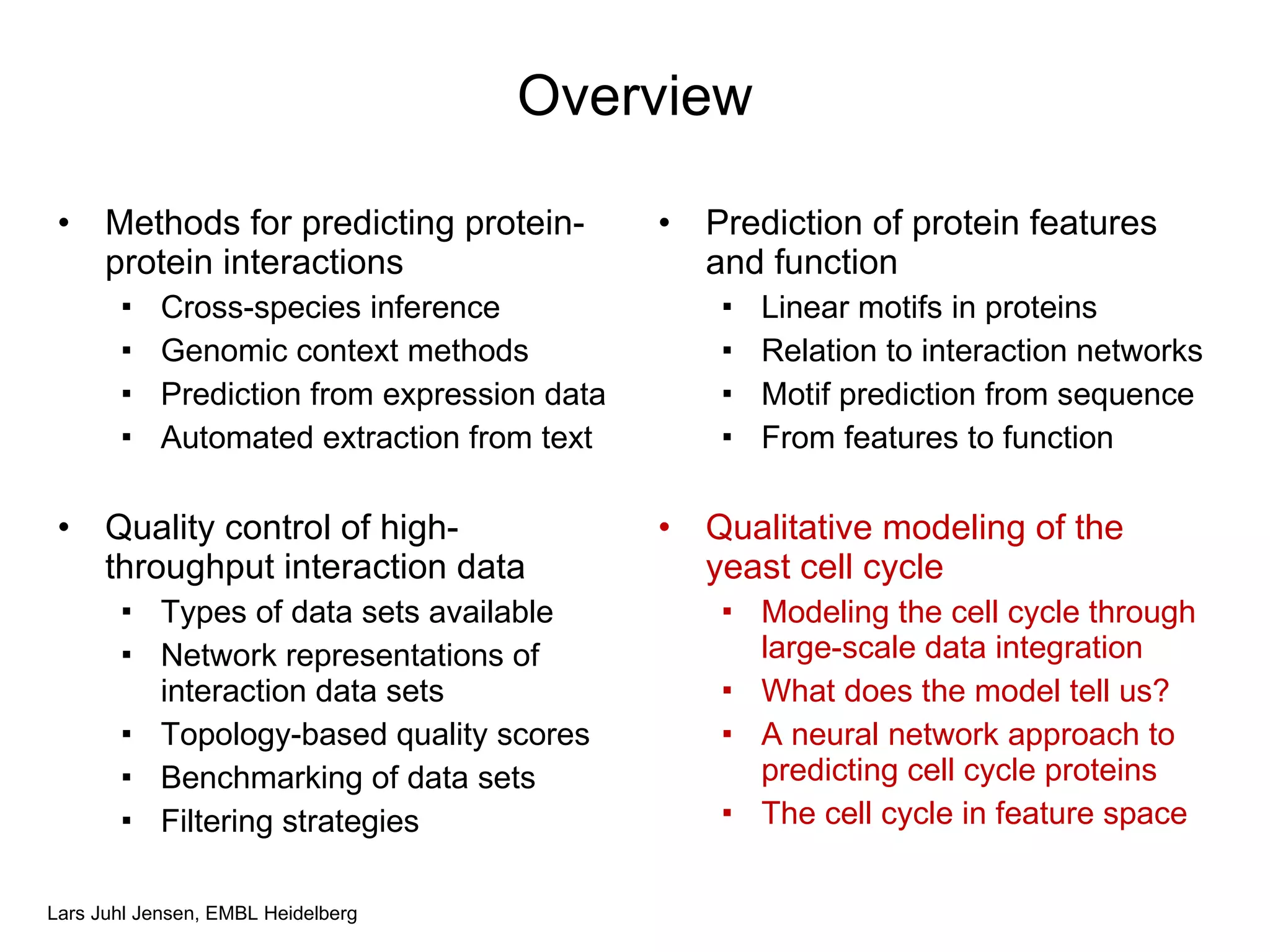
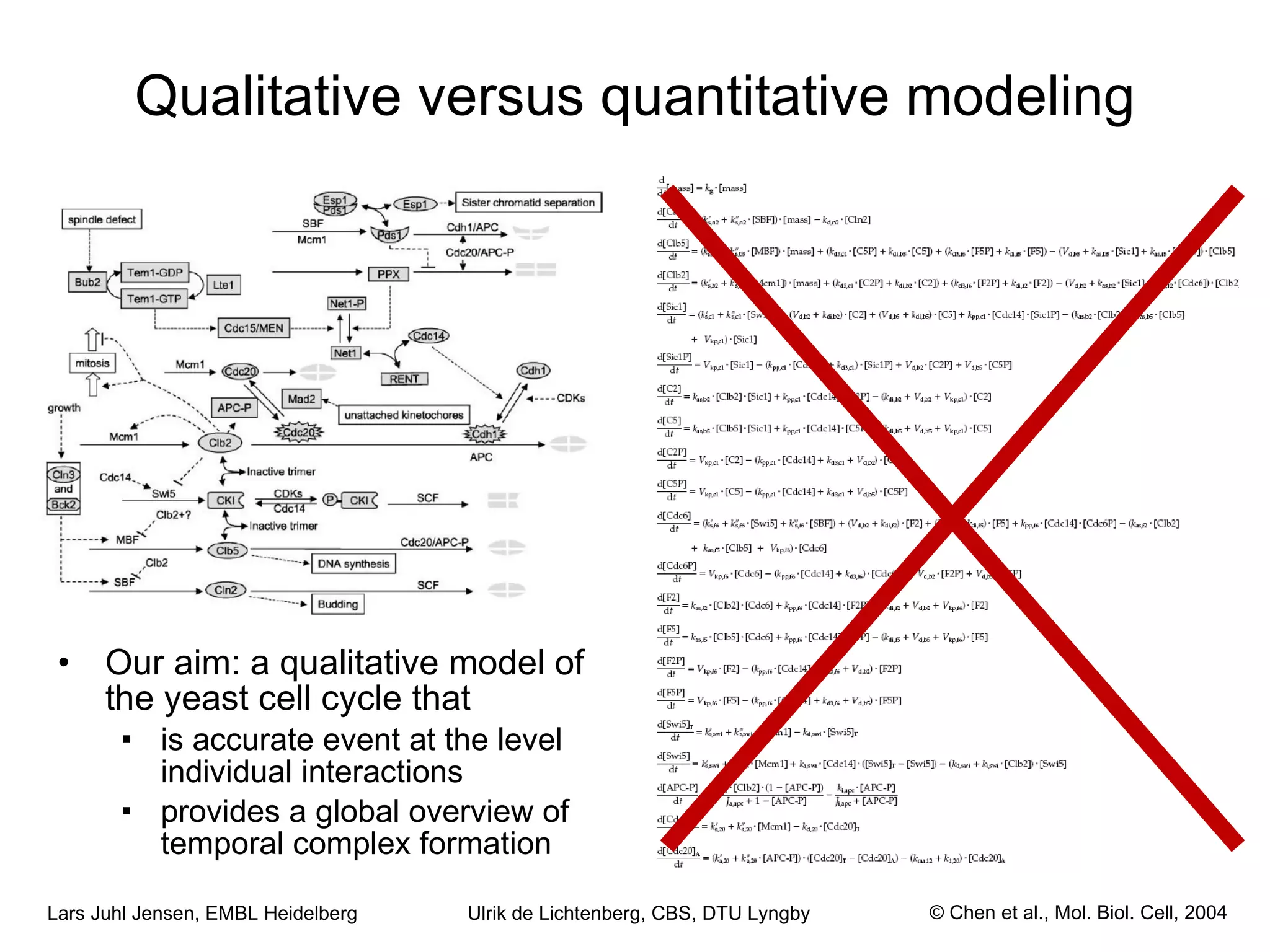
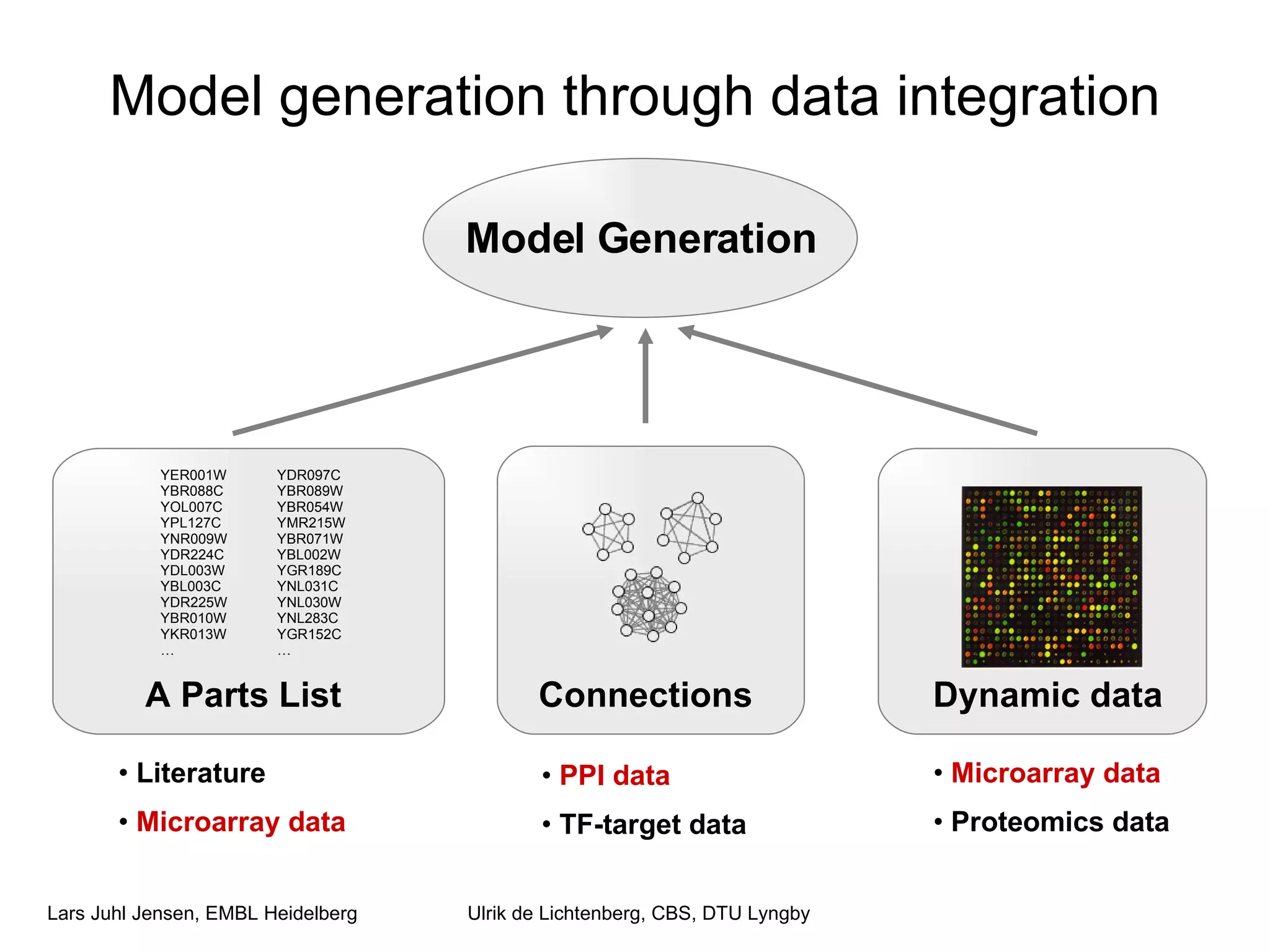
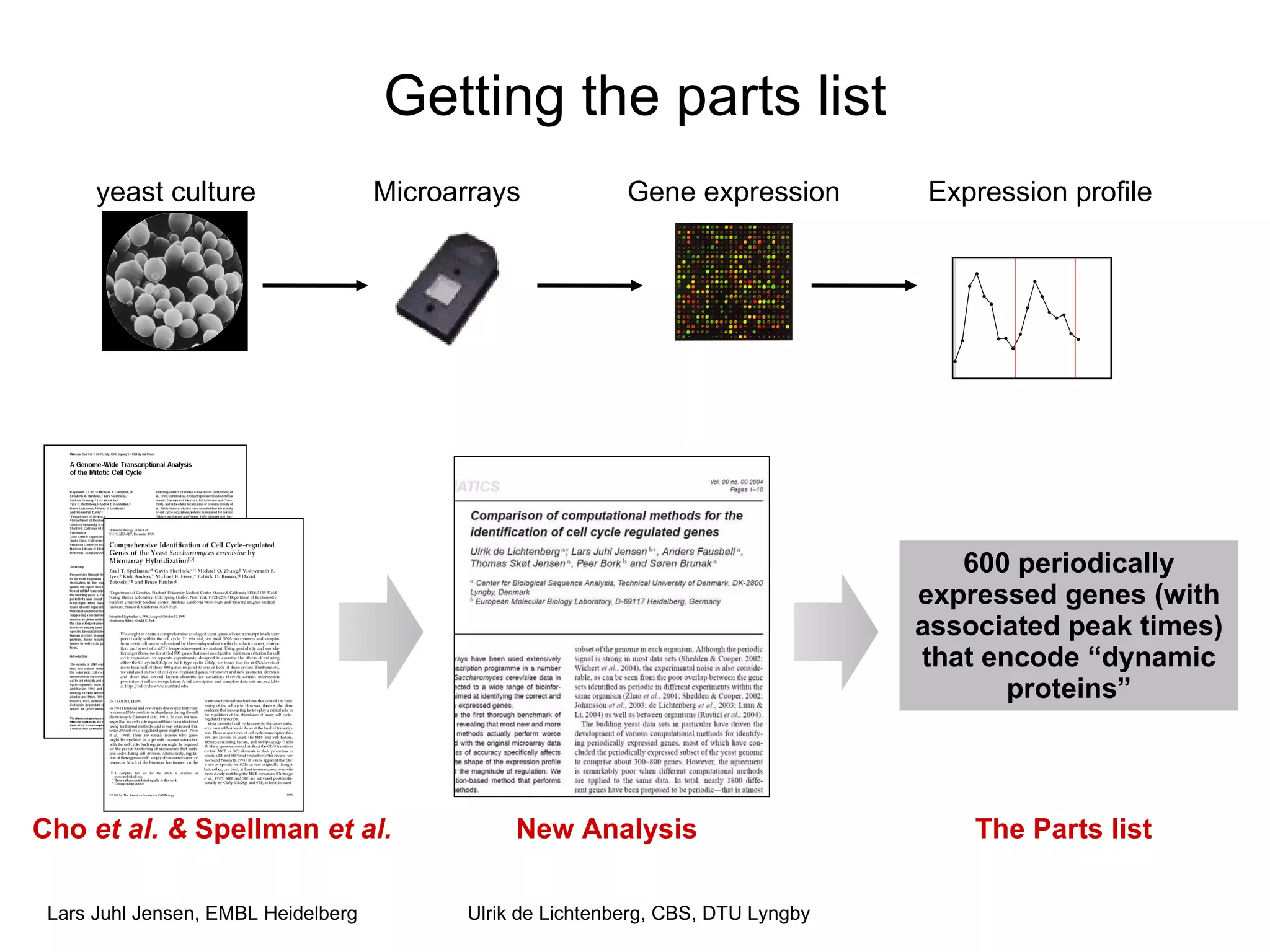
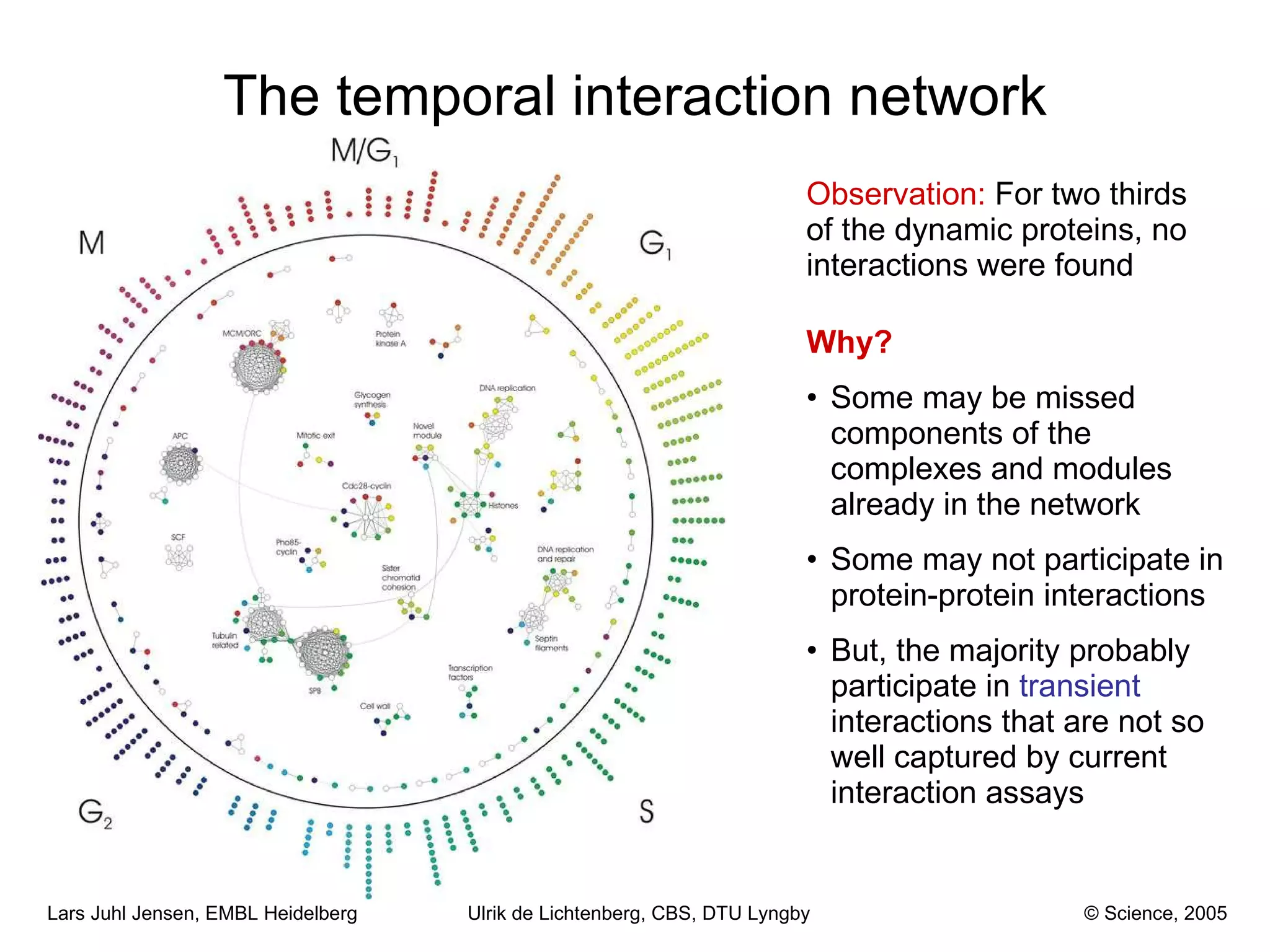
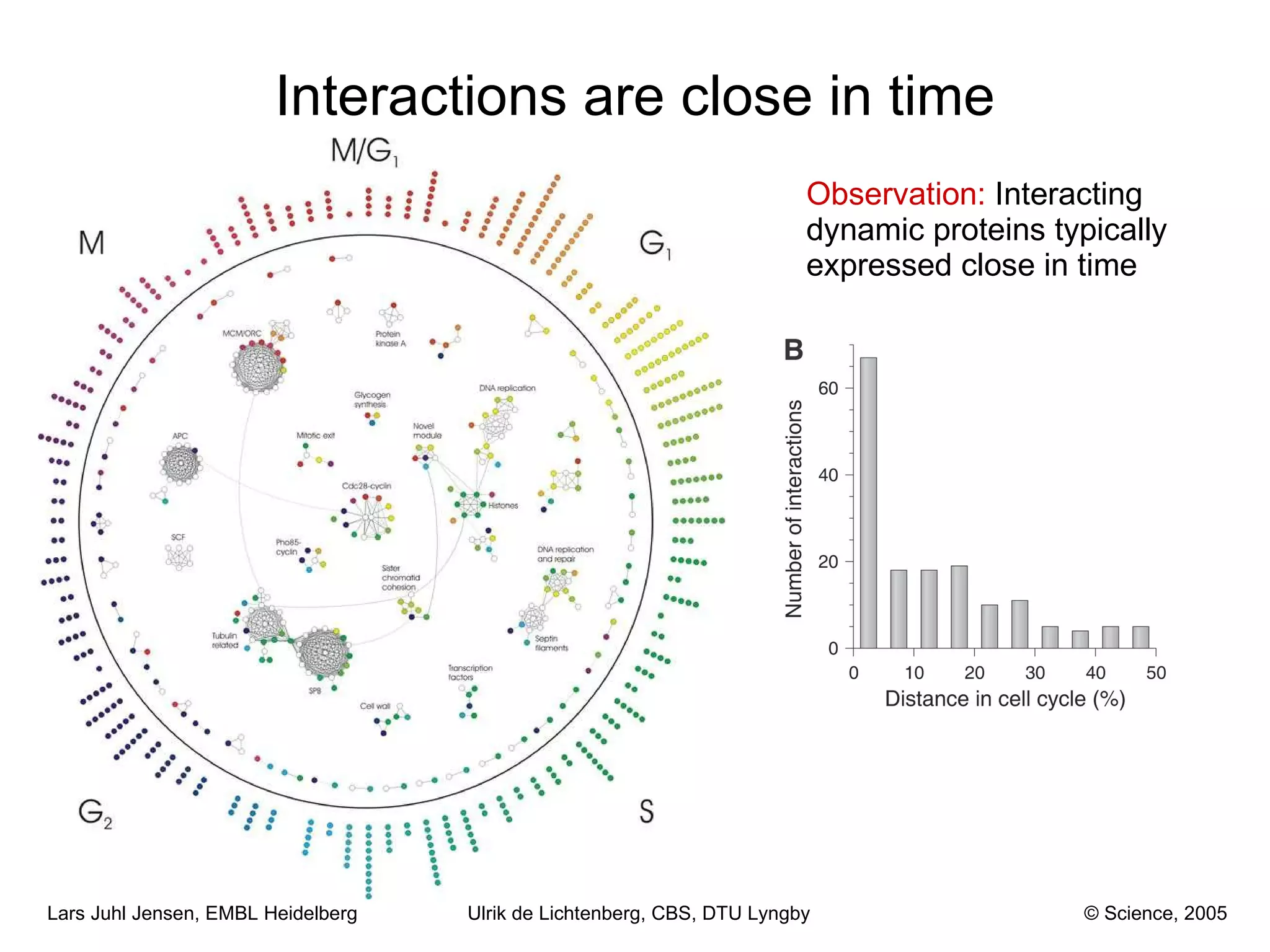
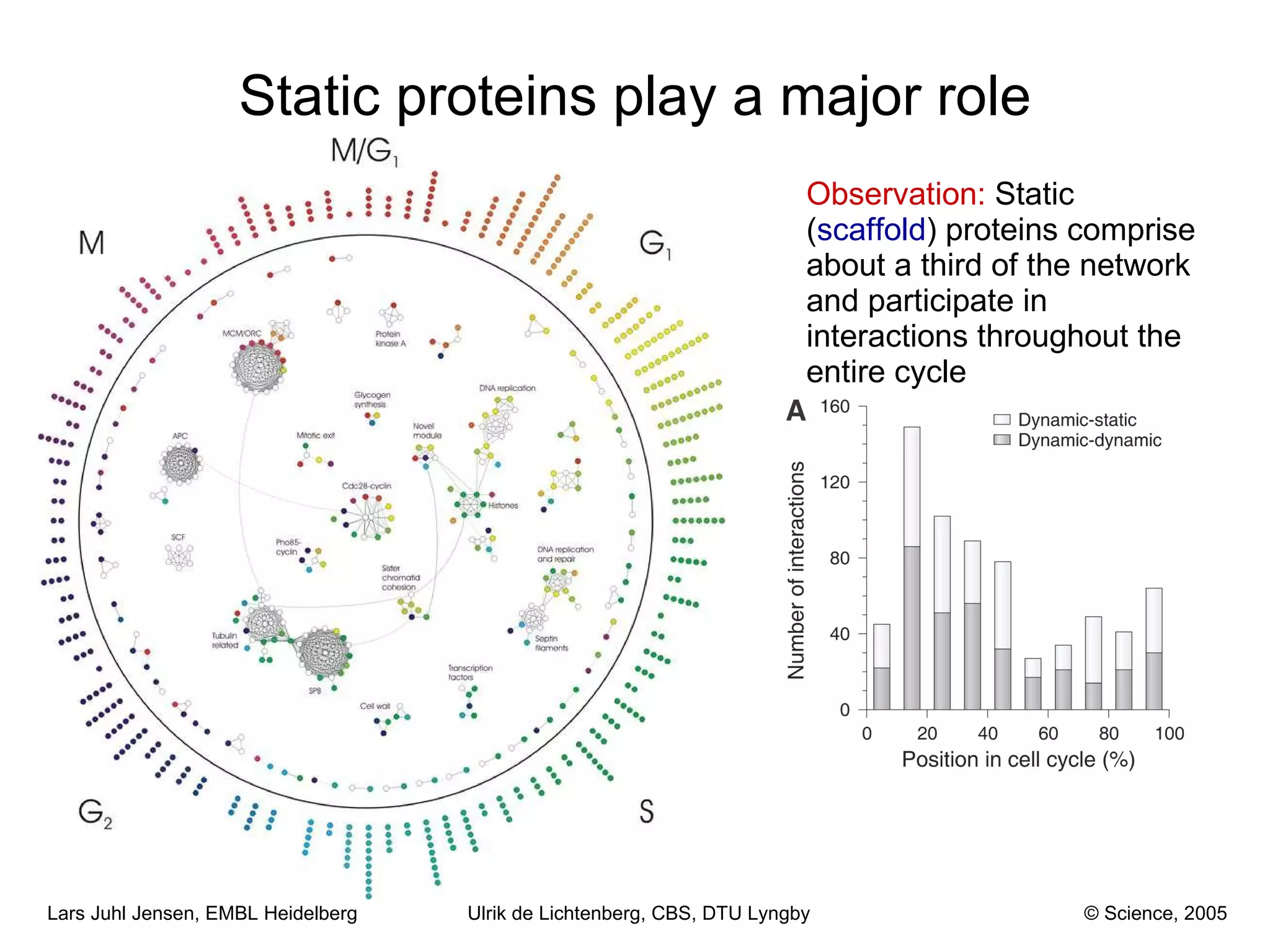
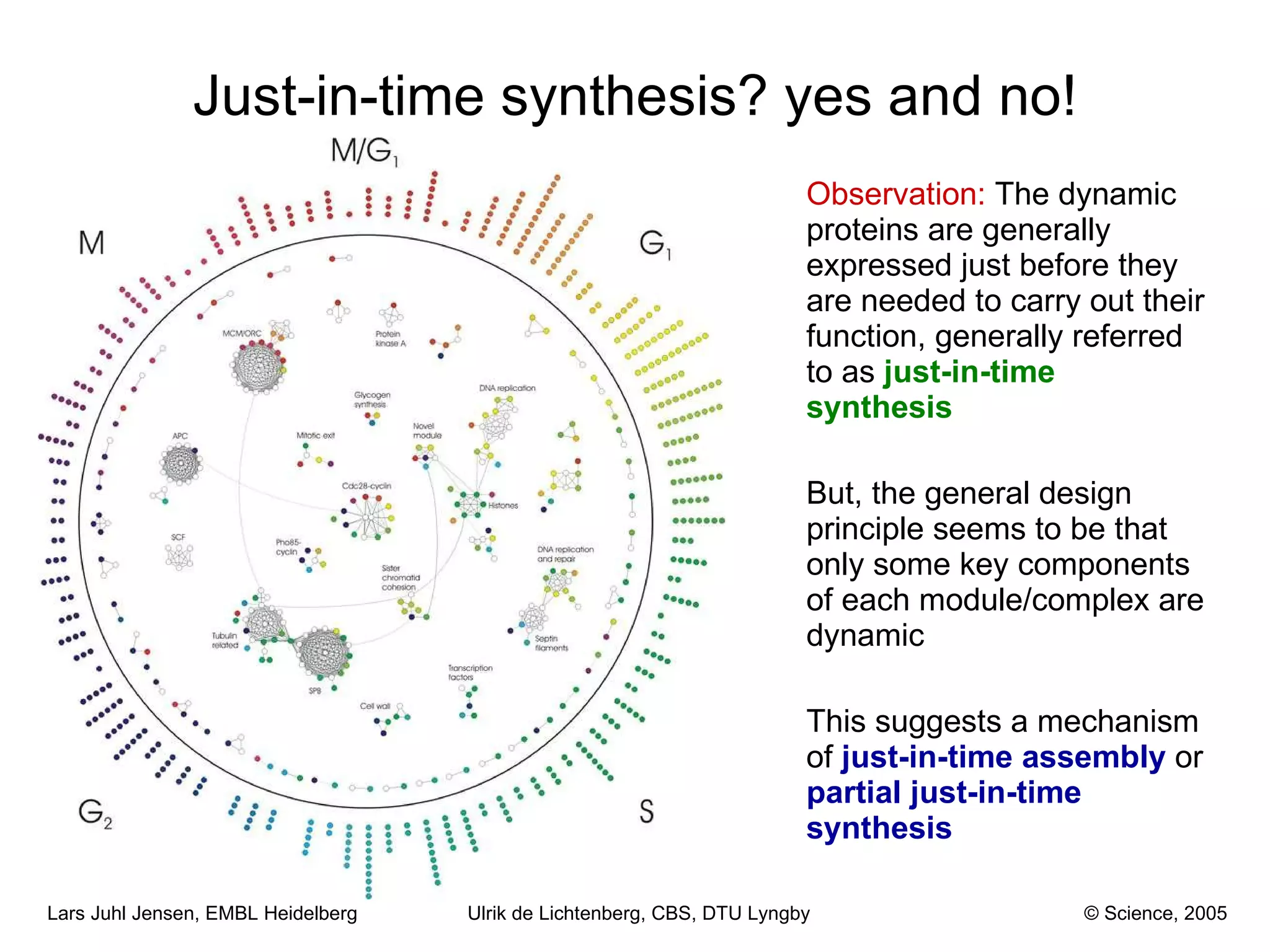
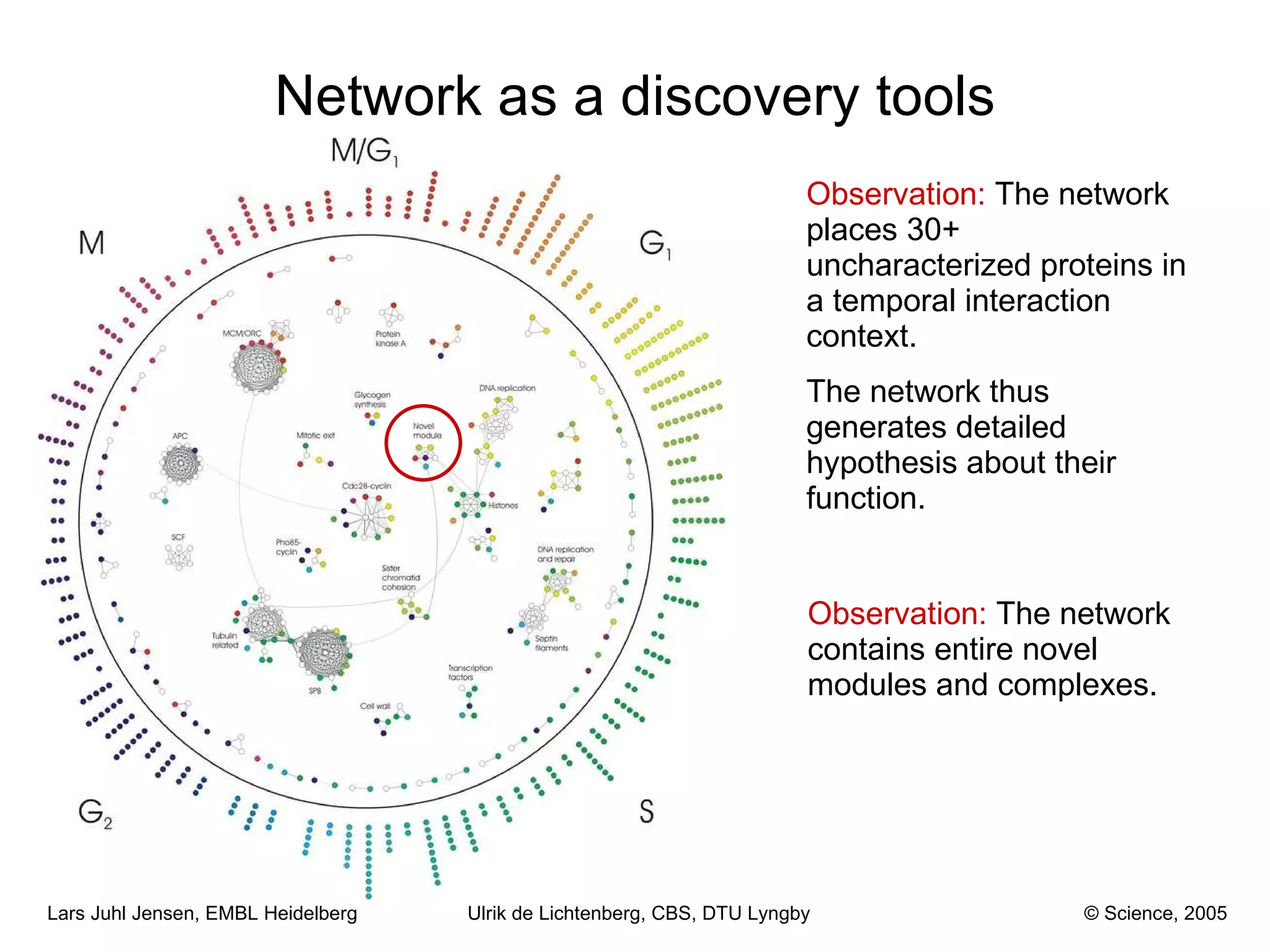
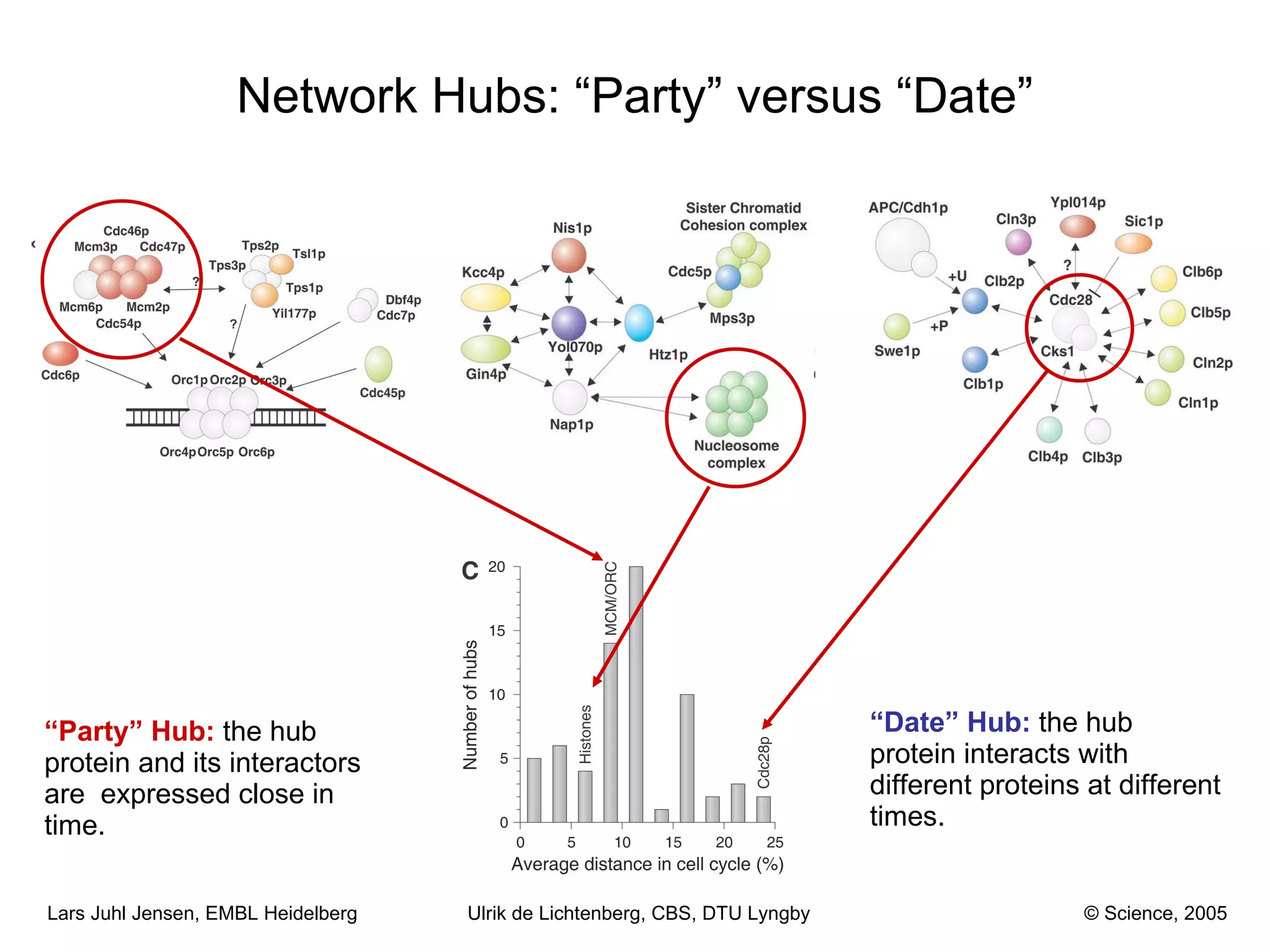
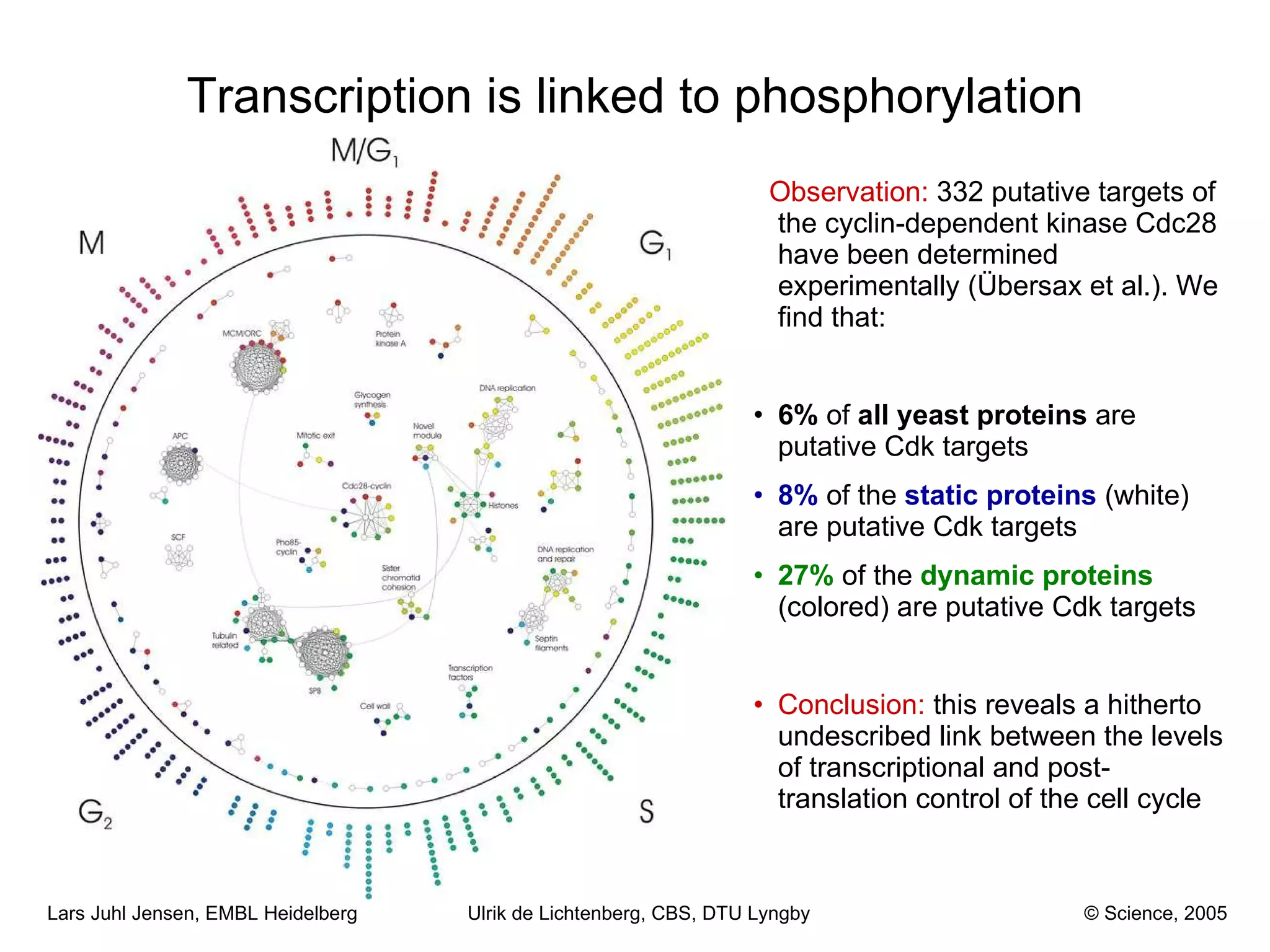
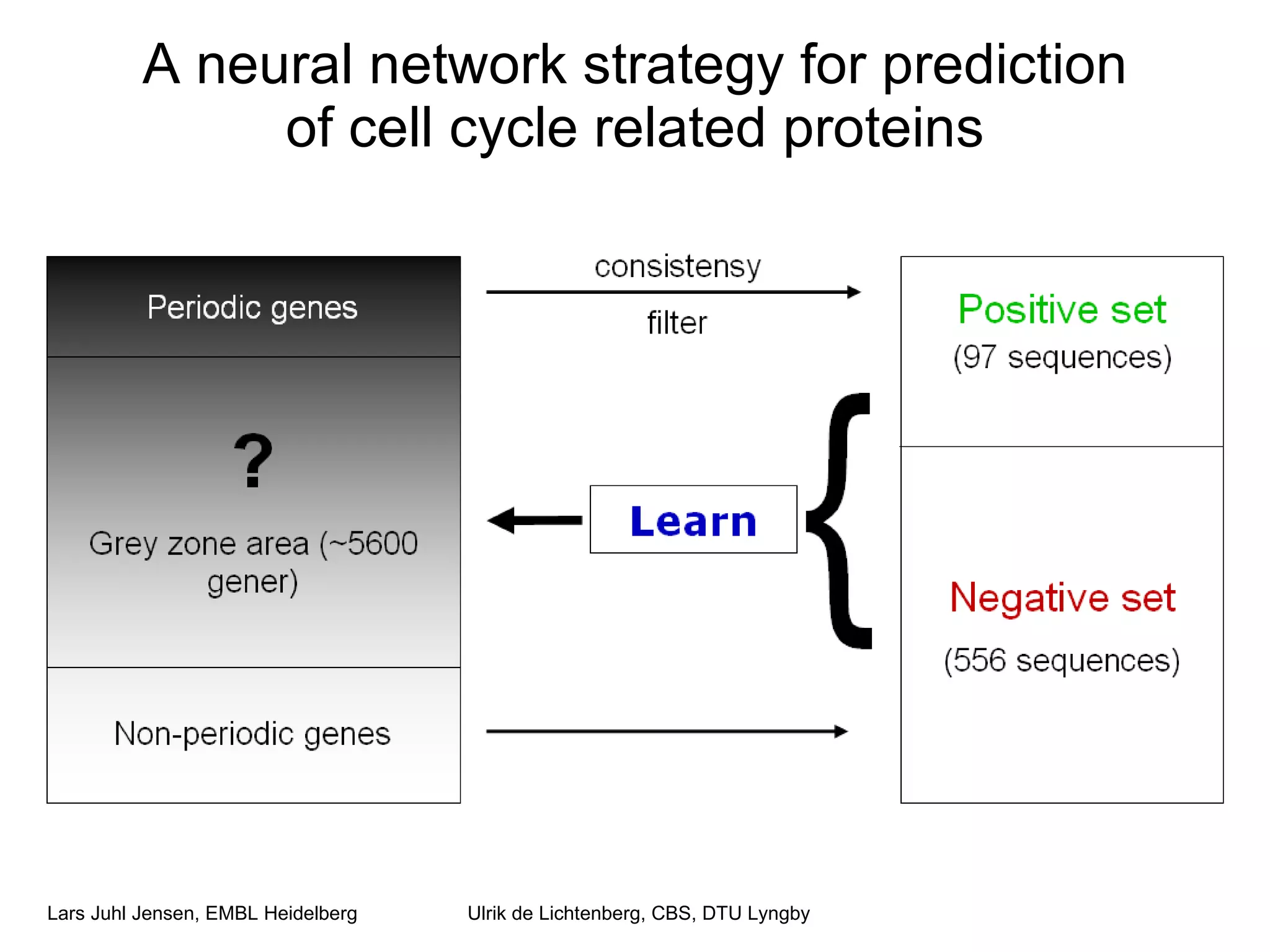
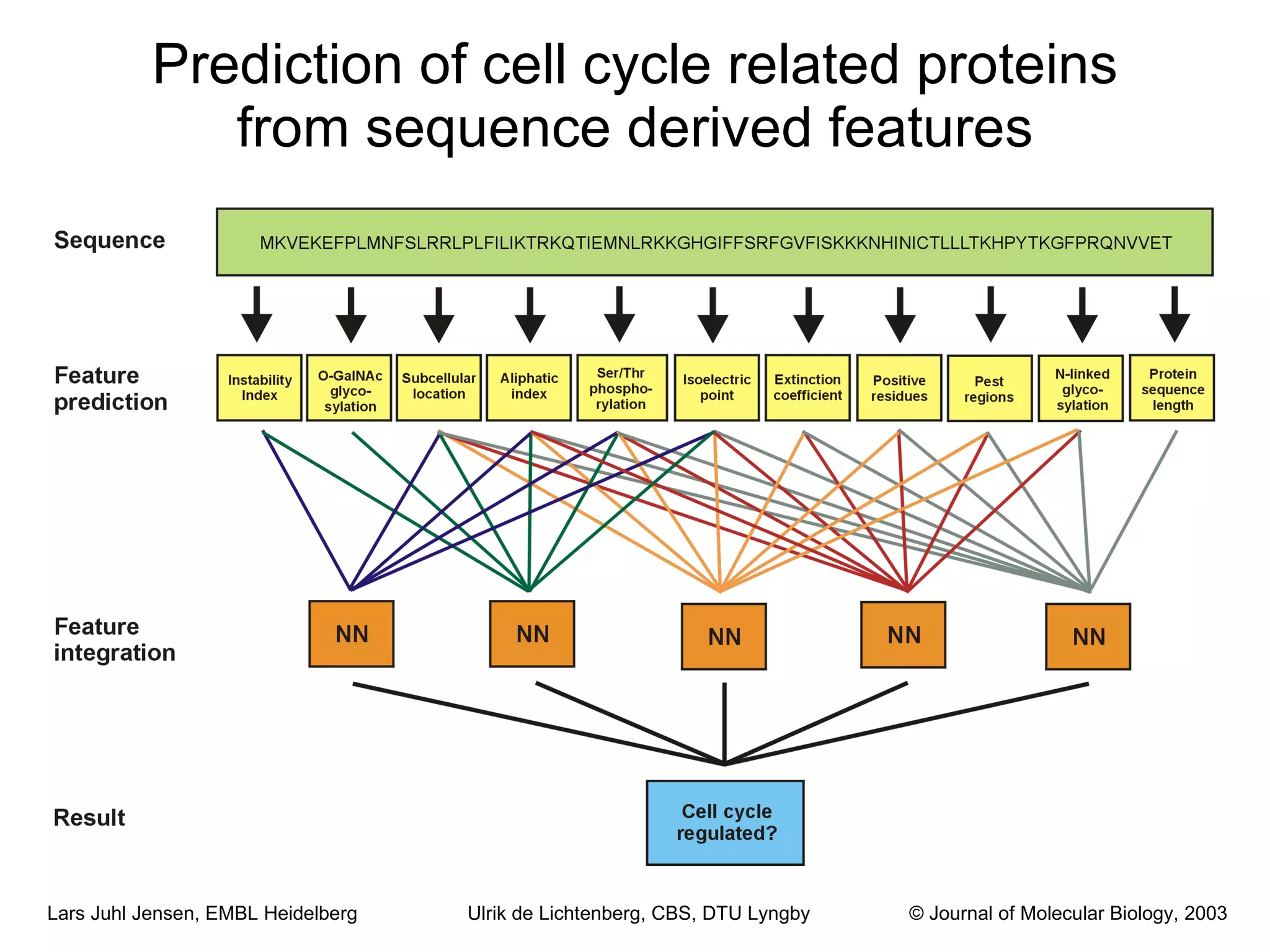
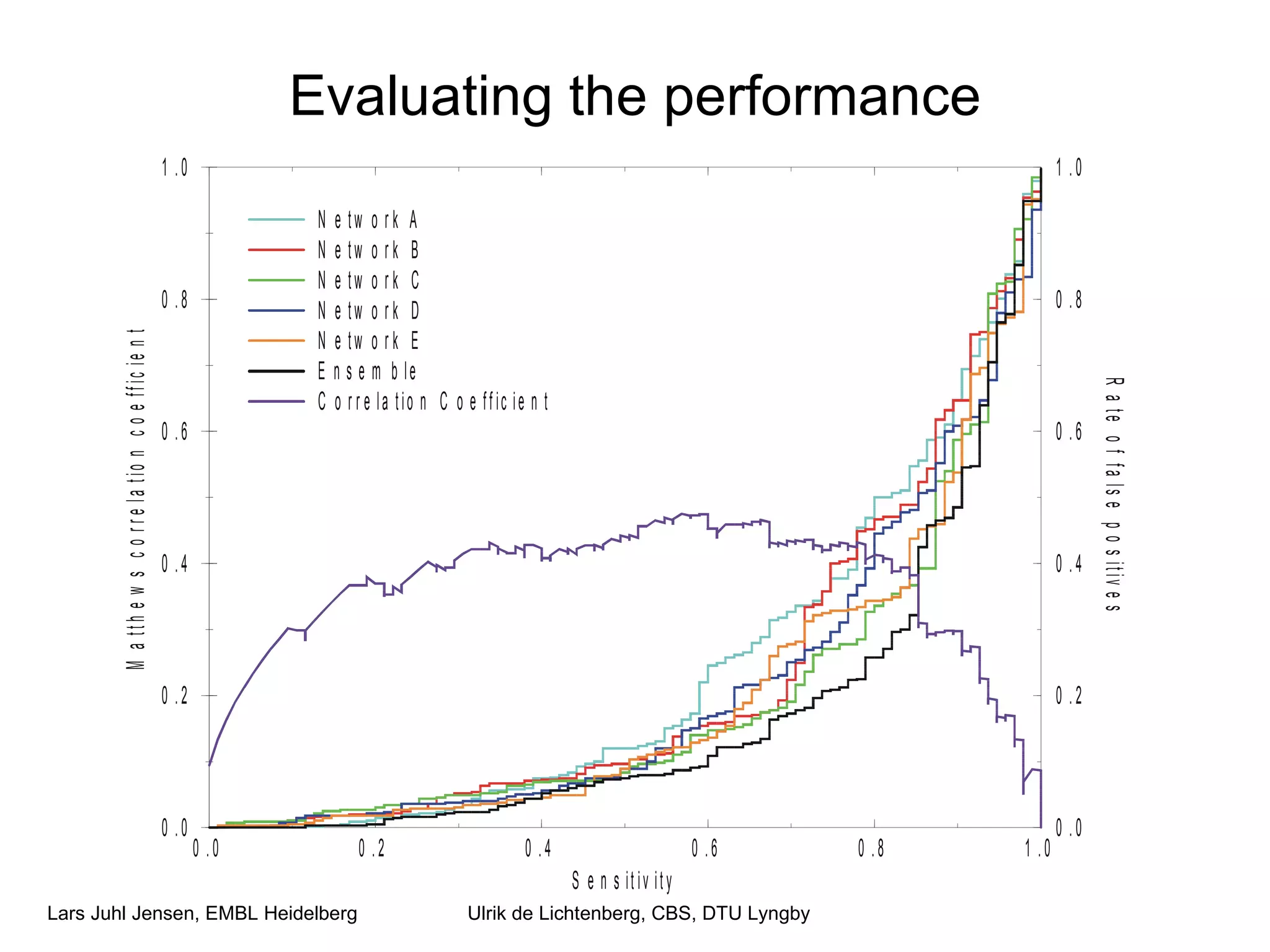
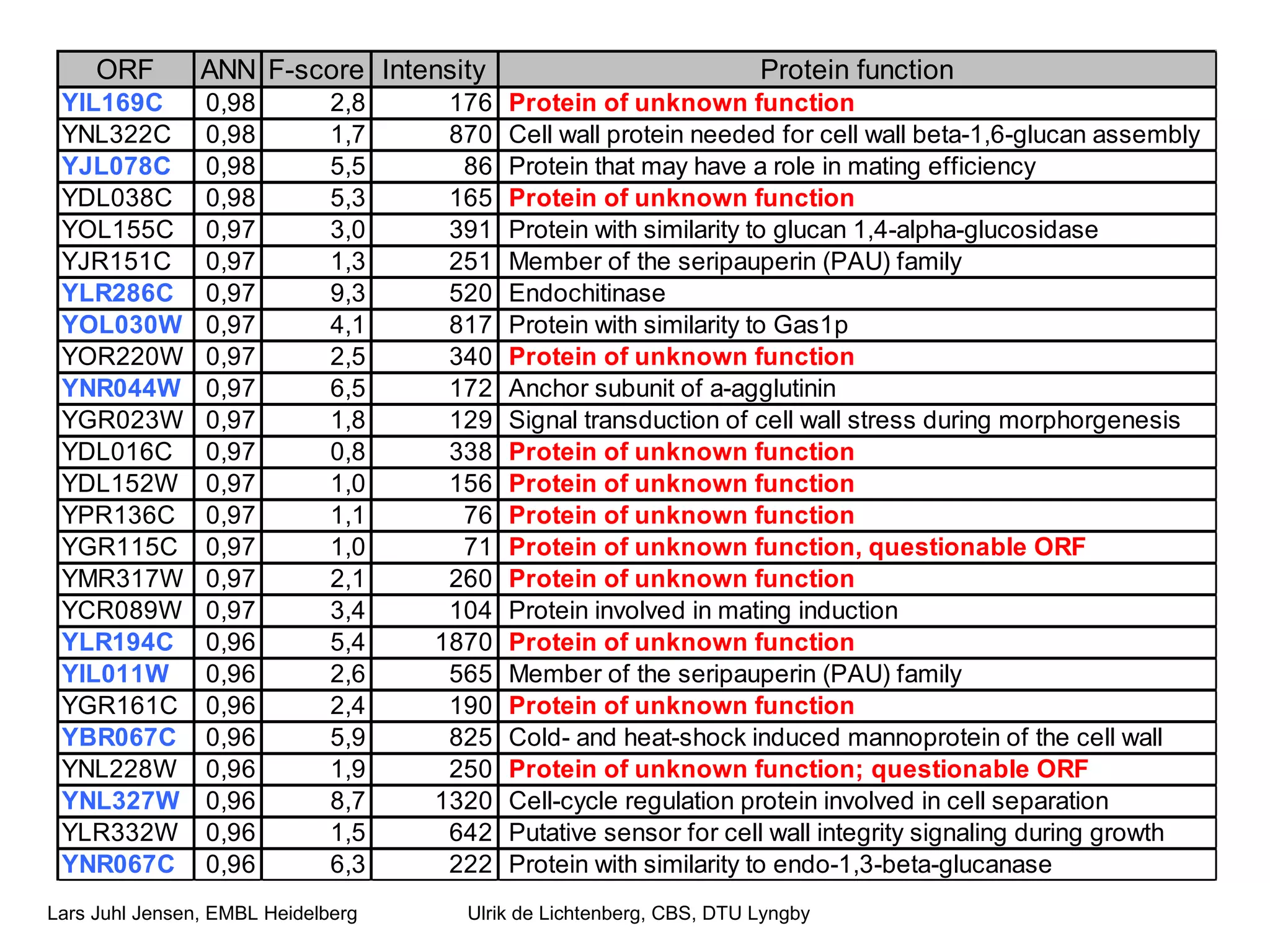
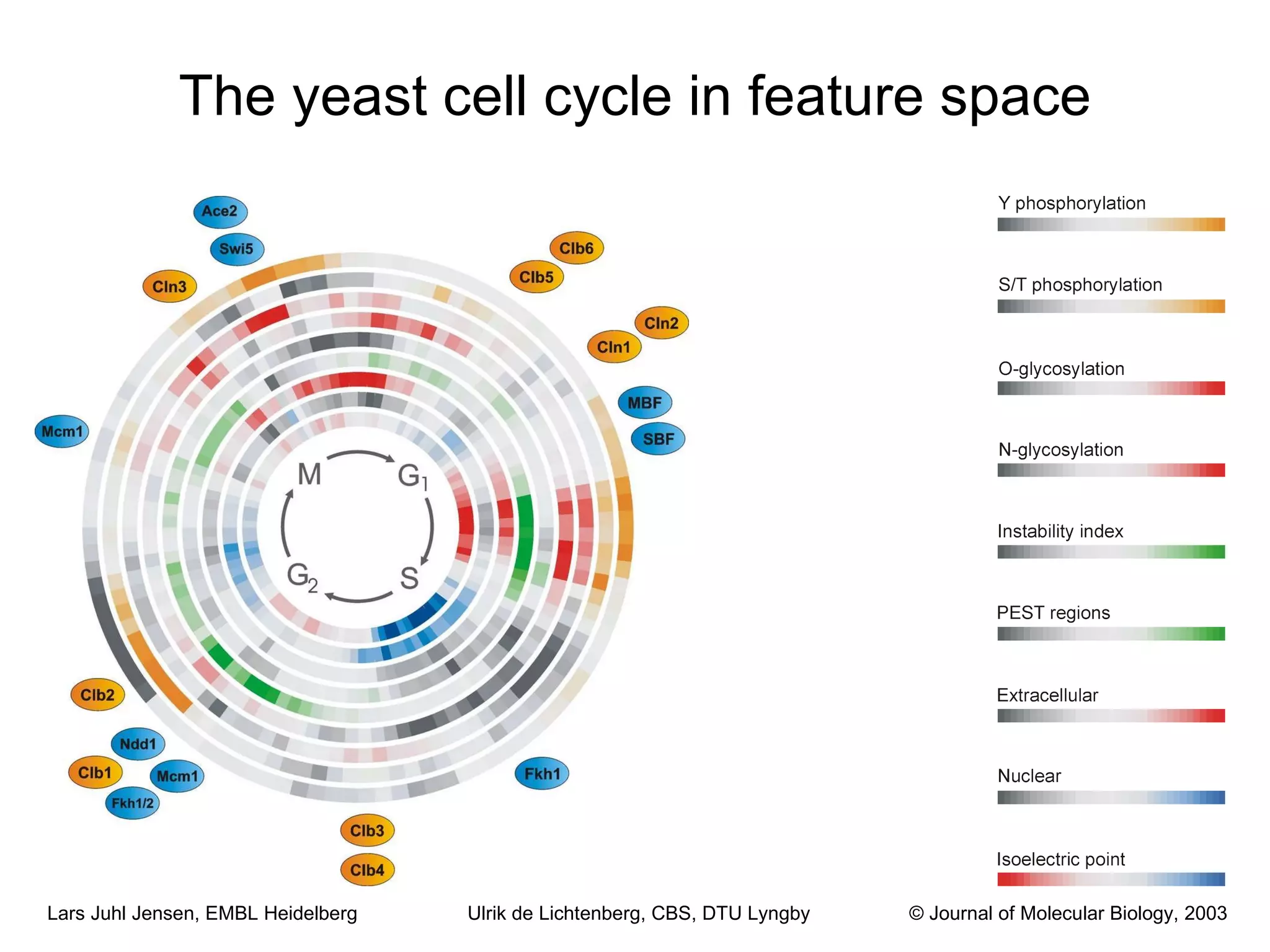
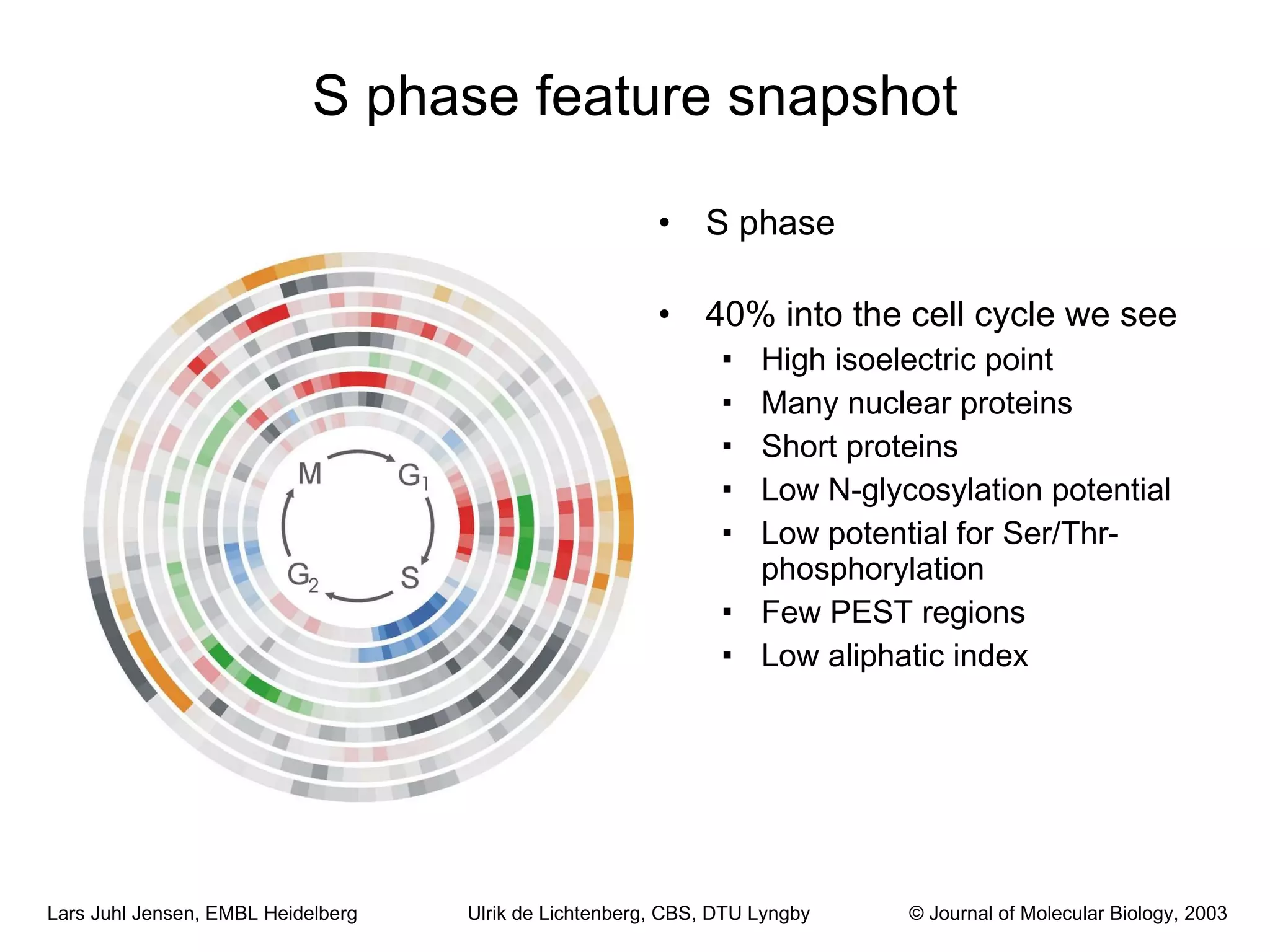
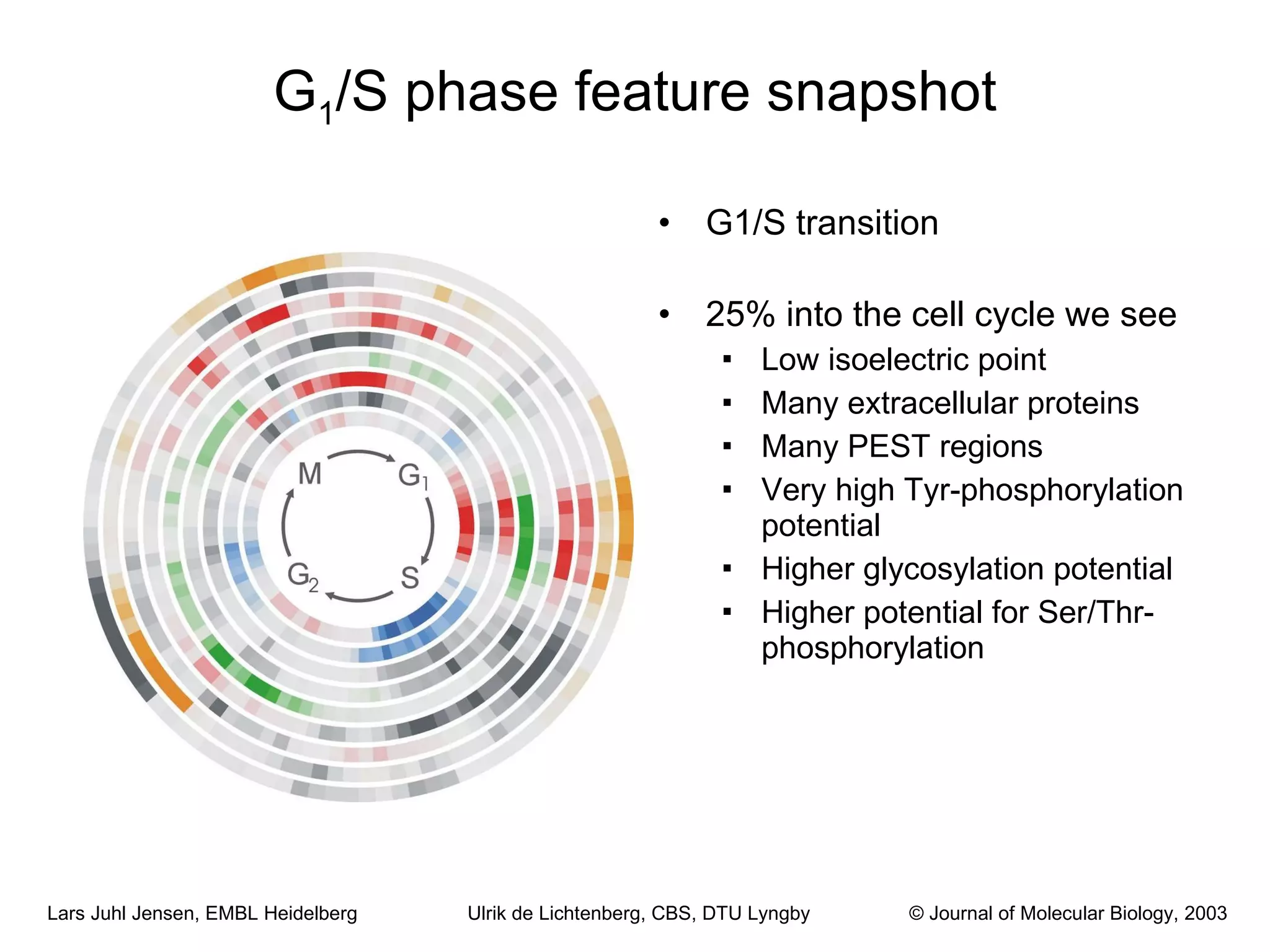
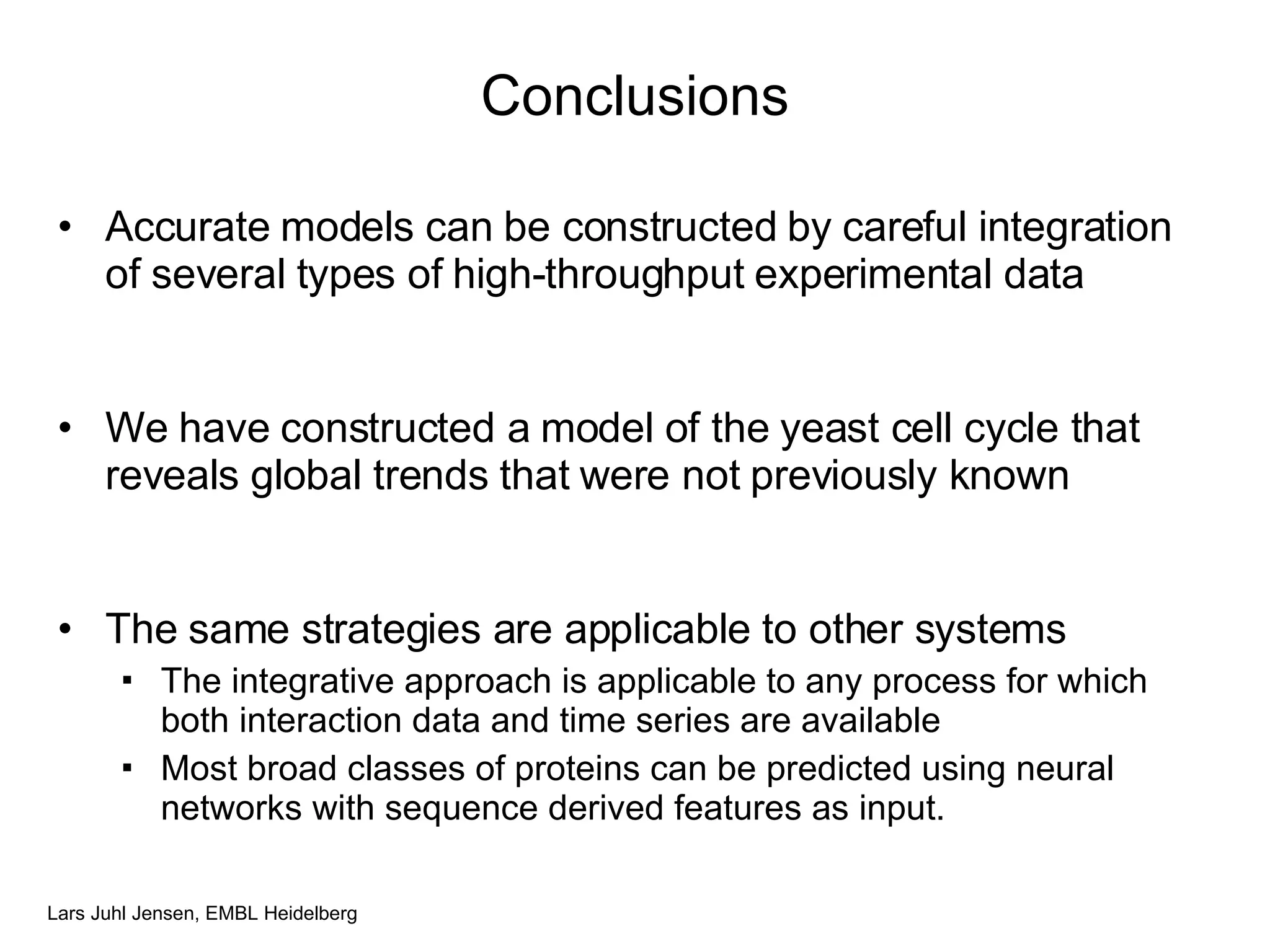
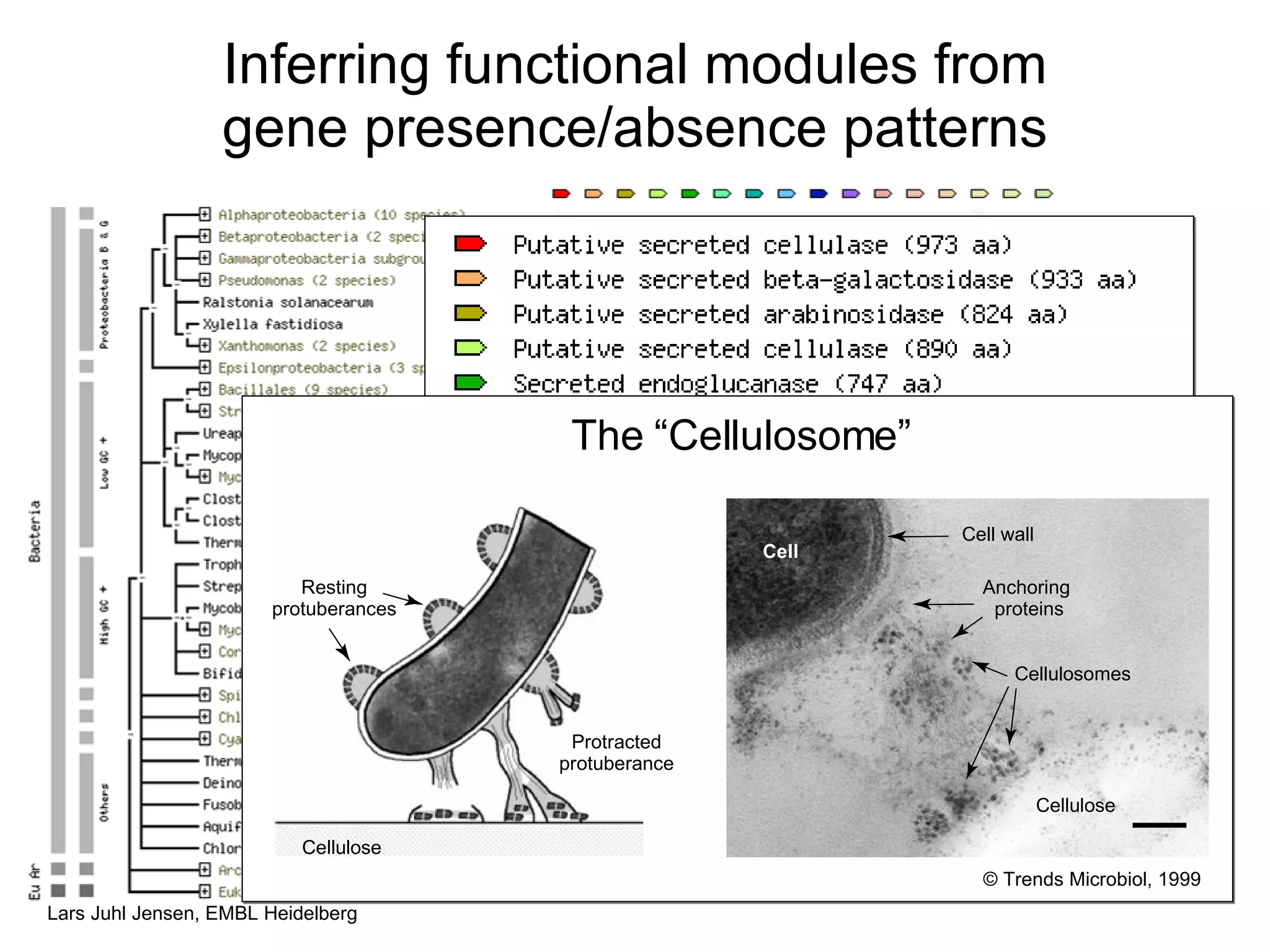
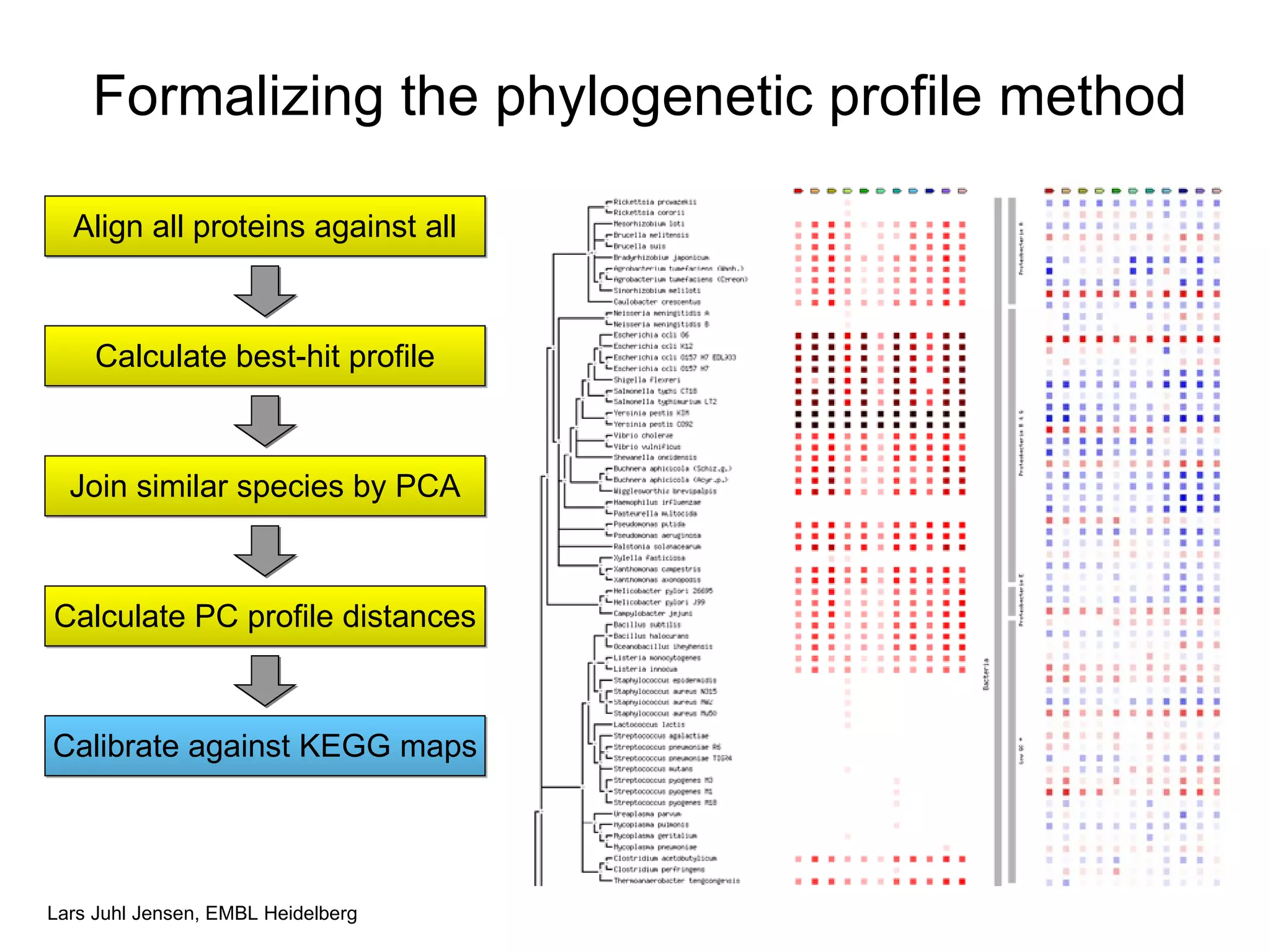
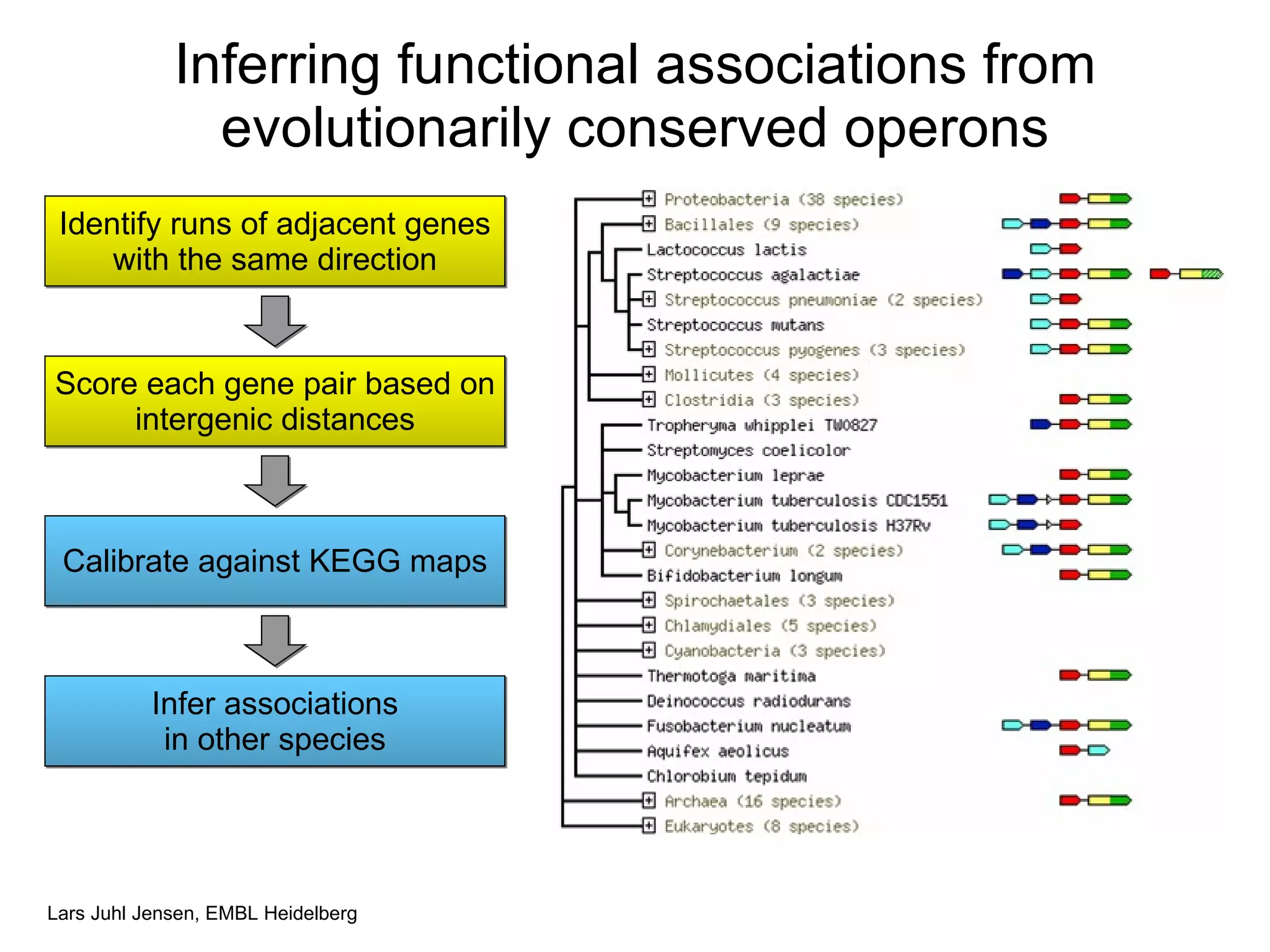
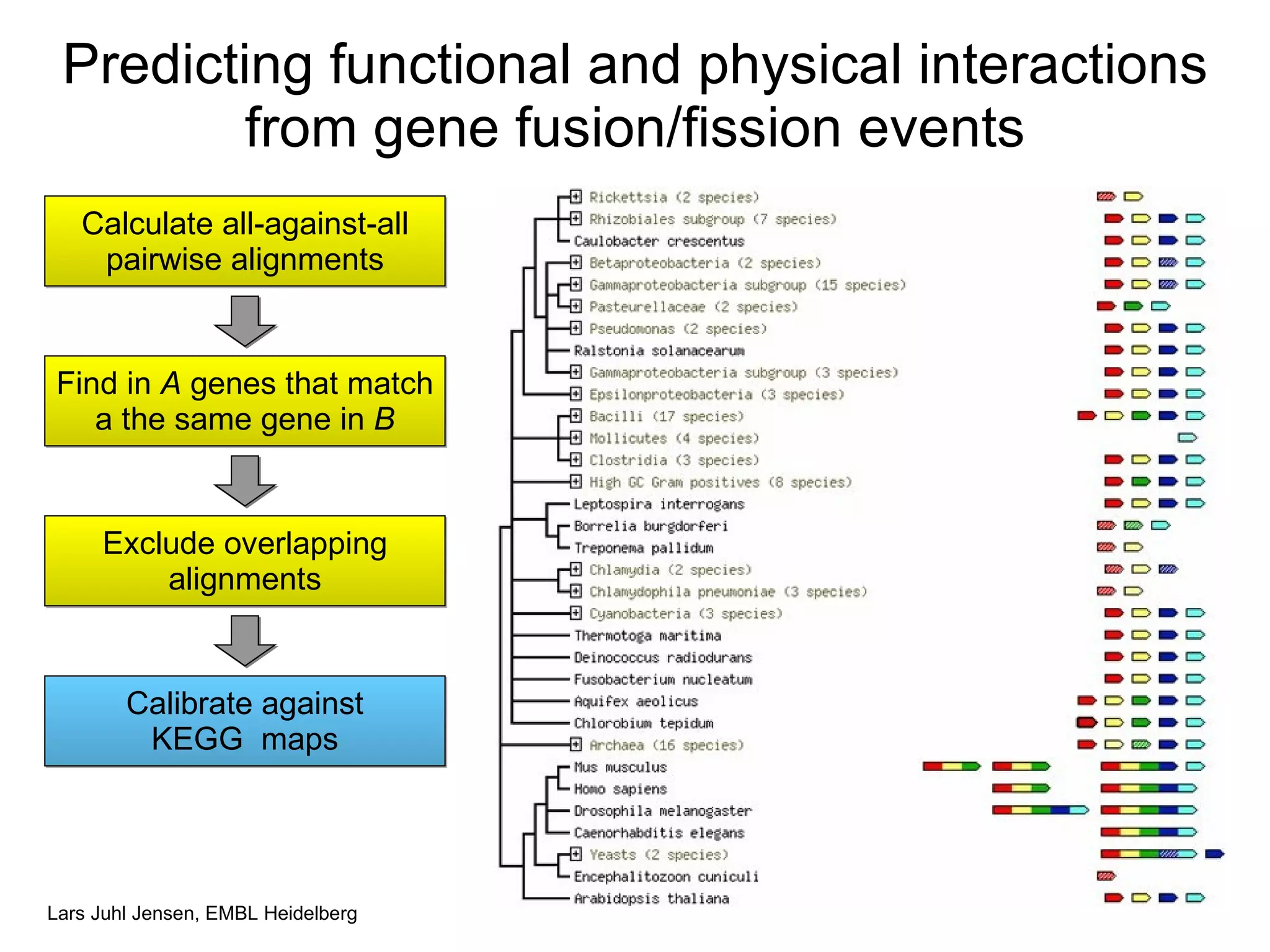
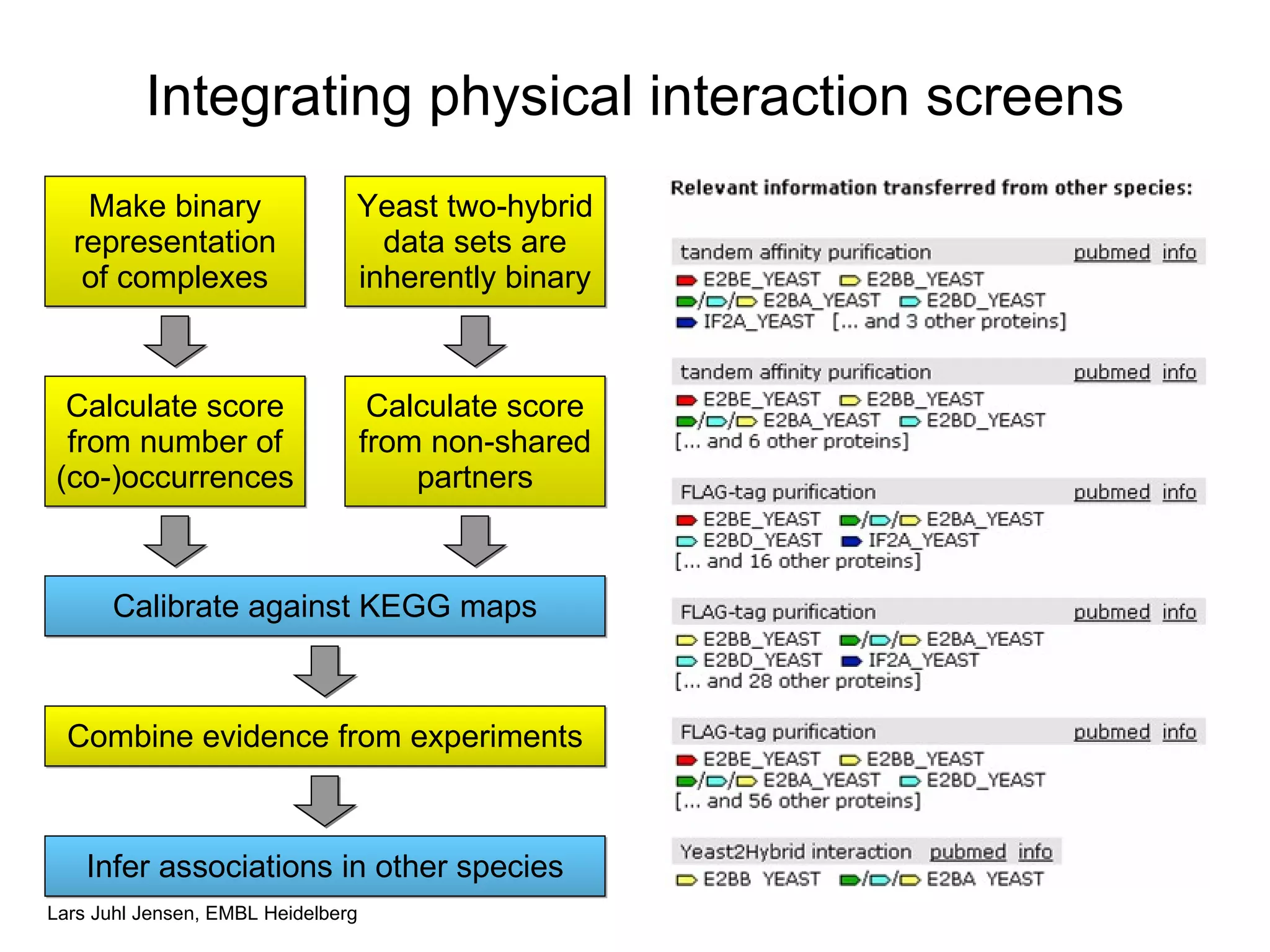
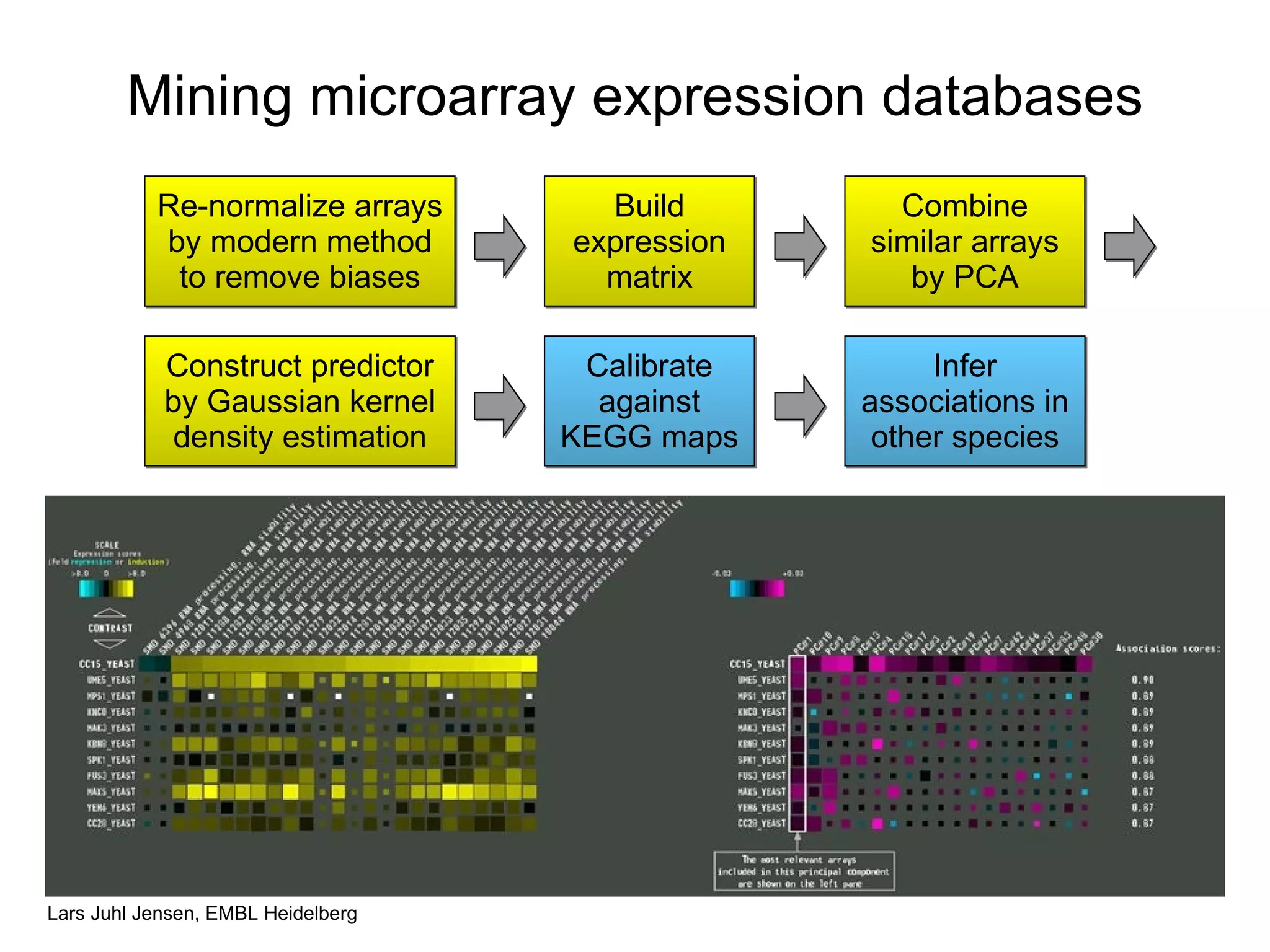
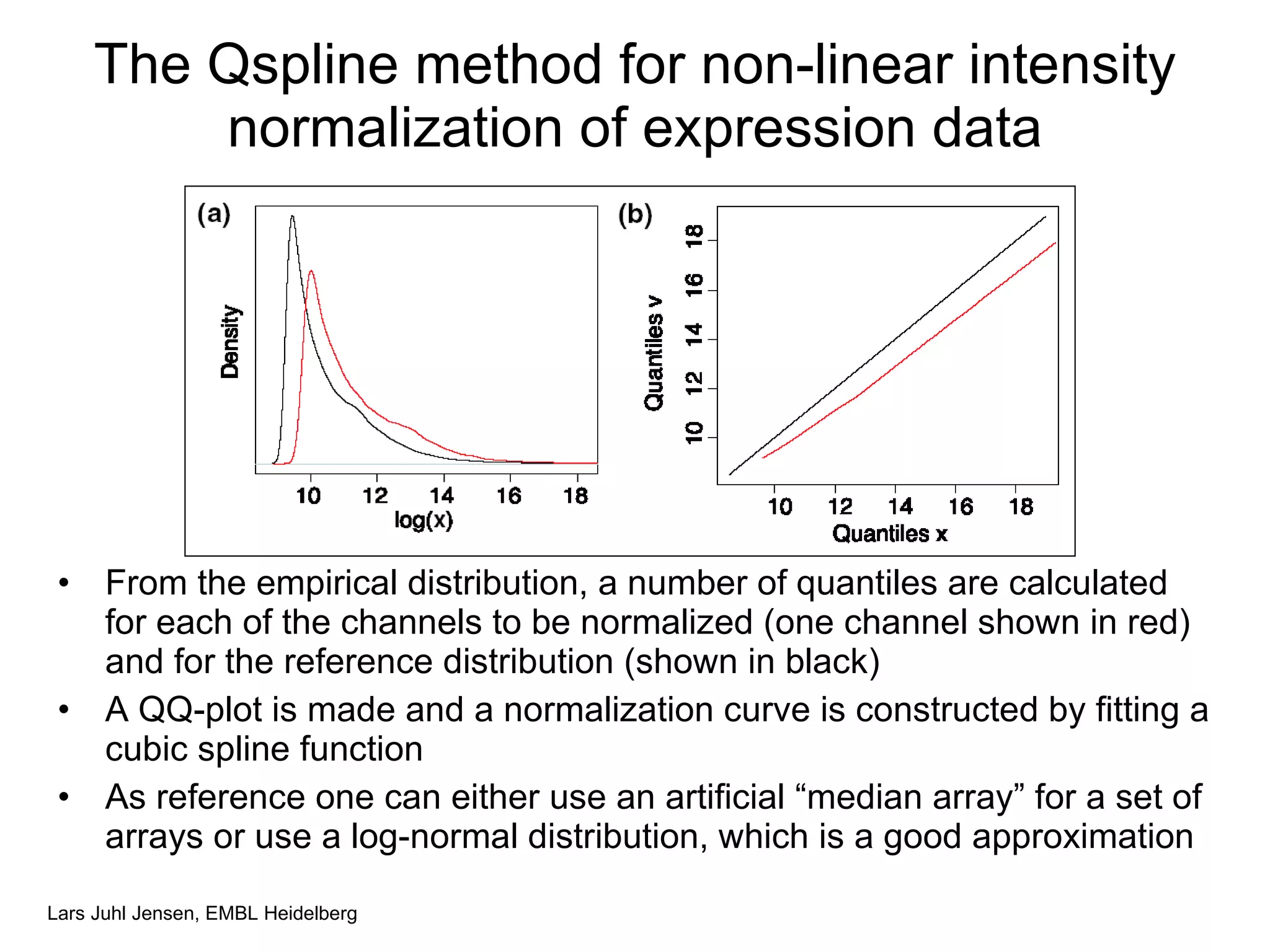
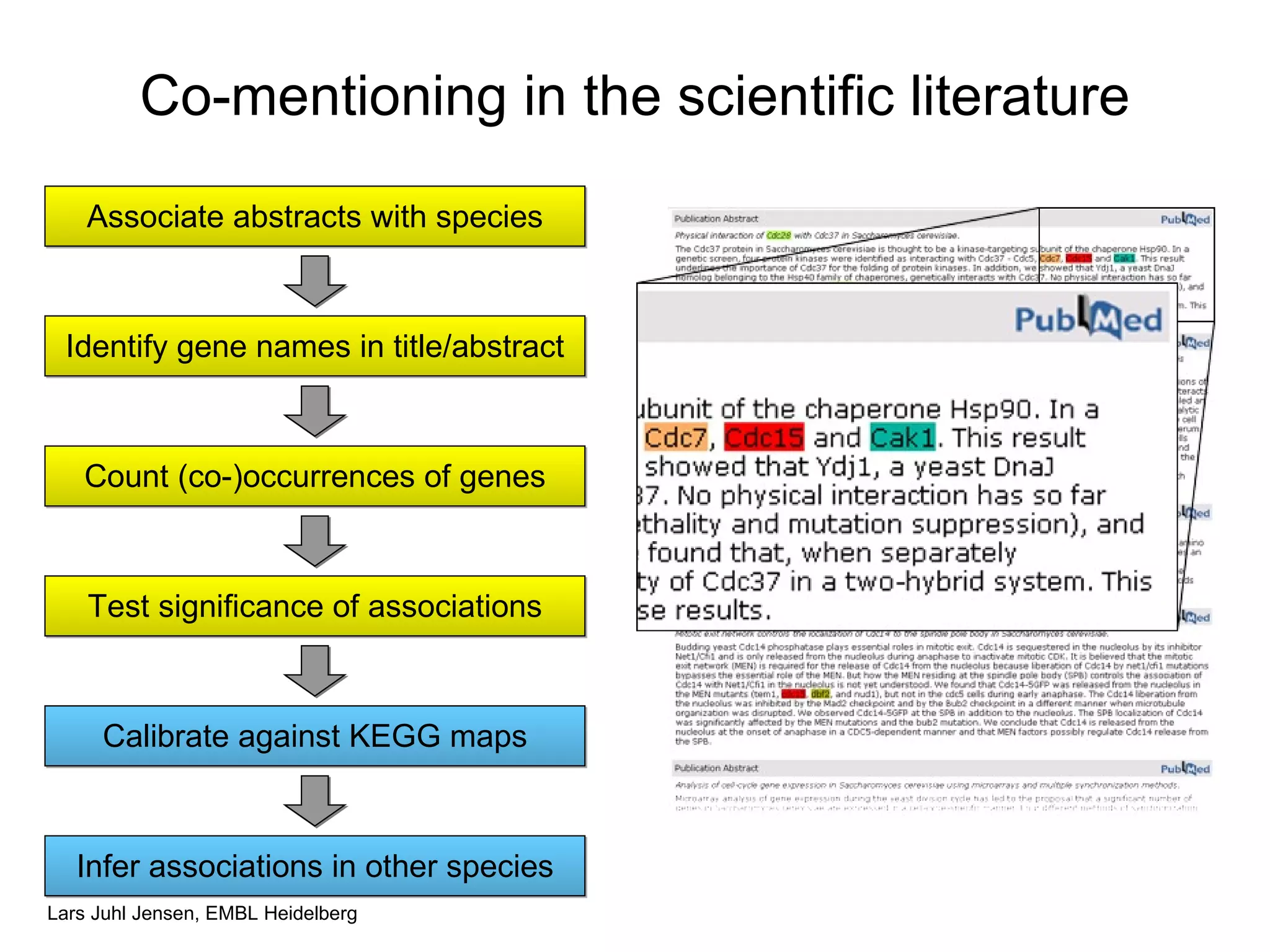
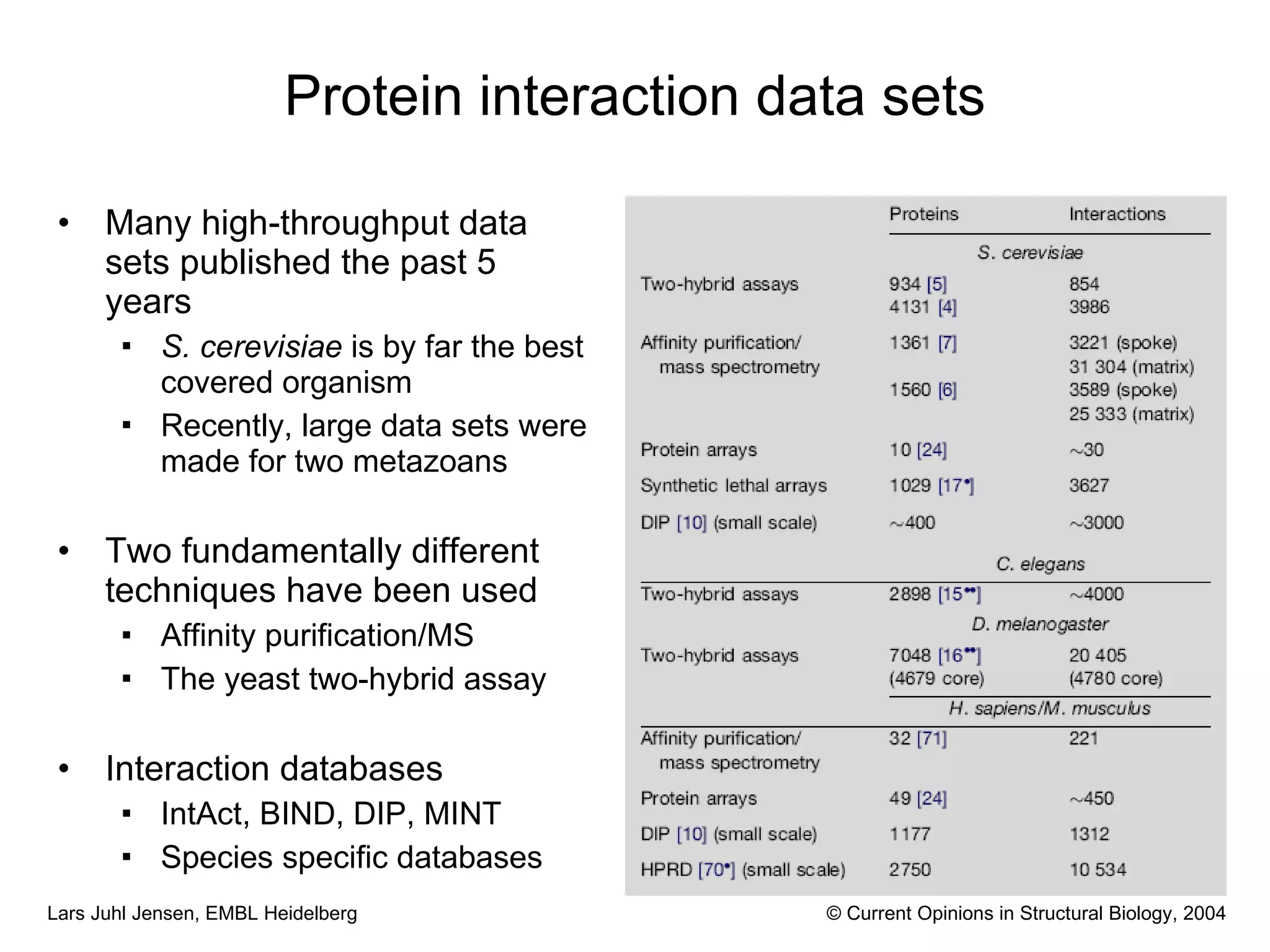
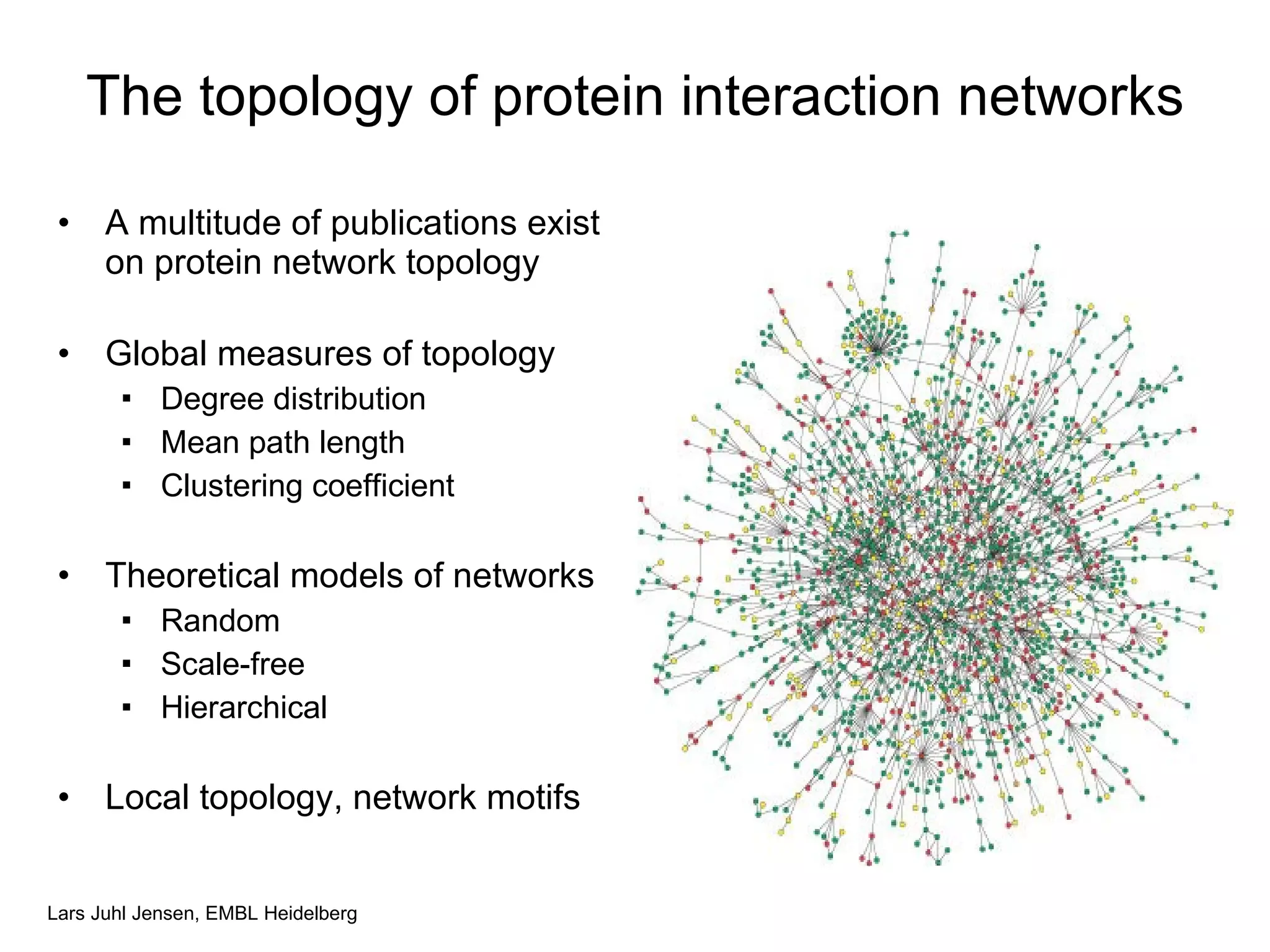
This document discusses various methods for integrating large-scale proteomics data to analyze protein-protein interaction networks, predict protein functions, and model biological processes like the yeast cell cycle. It describes combining different types of large datasets, such as genomic context, expression data, and text mining of literature, to infer functional relationships. Quality control methods are discussed for filtering high-throughput interaction datasets. The document also covers predicting protein features from sequence alone, like linear motifs and post-translational modifications, and relating these to interaction networks and functions.



![Topology based quality scores Scoring scheme for yeast two-hybrid data: S1 = -log((N 1 +1) · (N 2 +1)) N 1 and N 2 are the numbers of non-shared interaction partners Similar scoring schemes have been published by Saito et al. Scoring scheme for complex pull-down data: S2 = log[(N 12 · N)/((N 1 +1) · (N 2 +1))] N 12 is the number of purifications containing both proteins N 1 is the number containing protein 1, N 2 is defined similarly N is the total number of purifications Both schemes aim at identifying ubiquitous interactors](https://image.slidesharecdn.com/jen05talk1-1217966787644673-8/75/Proteomics-Analysis-and-integration-of-large-scale-data-sets-35-2048.jpg)


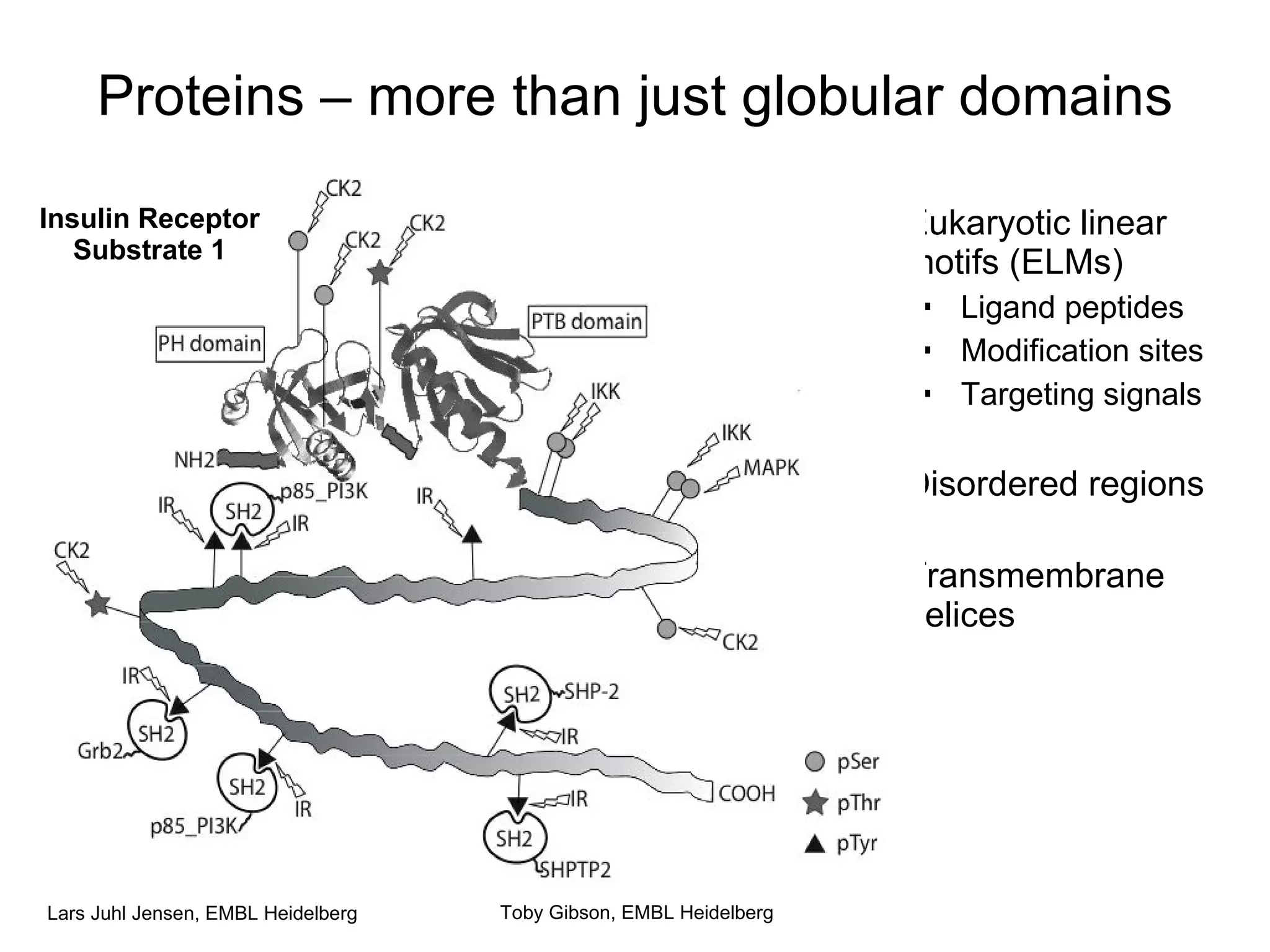
![Most ELMs are “information poor” Weak/short consensus sequences for ELMs The typical ELM only has three conserved residues Some variance is often allowed even for these ELMs are very hard to predict from sequence Consensus sequences simply match everywhere The information is not in the local sequence Most ELMs can only be predicted using context Toby Gibson, EMBL Heidelberg L . C . E RB interaction [RK] .{0,1} V . F PP1 interaction R . L .{0,1} [FLIMVP] Cyclin binding motif SP . [KR] CDK phosphorylation L . . LL NR Box P . L . P MYND finger interaction F . . . W . . [LIV] MDM2-binding RGD Integrin-binding SKL$ Peroxisome targeting [RK][RK] . [ST] PKA phosphorylation](https://image.slidesharecdn.com/jen05talk1-1217966787644673-8/75/Proteomics-Analysis-and-integration-of-large-scale-data-sets-47-2048.jpg)