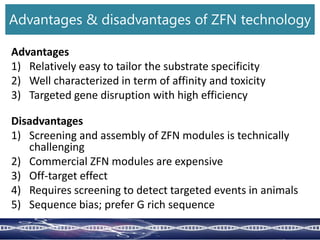
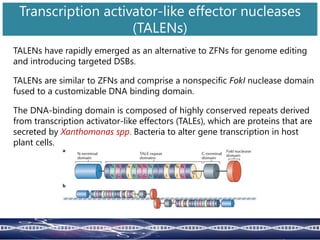
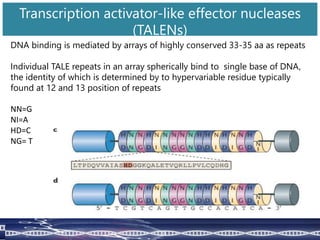
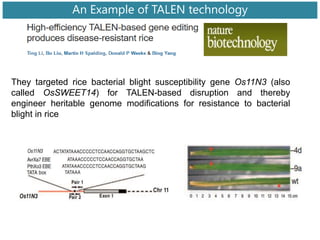
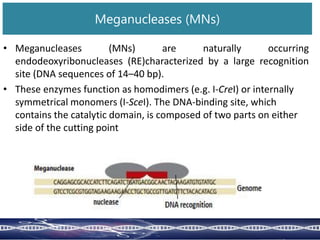
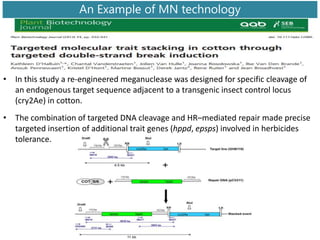
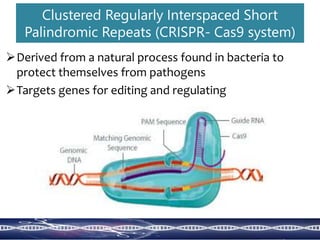
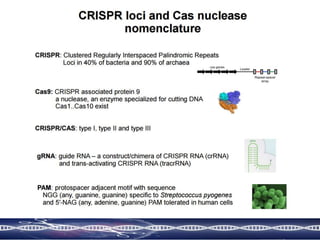
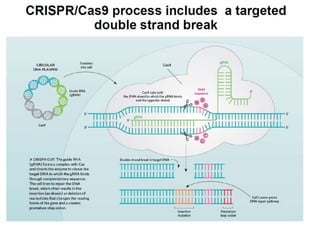
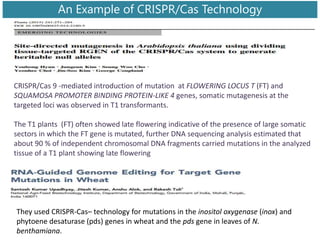
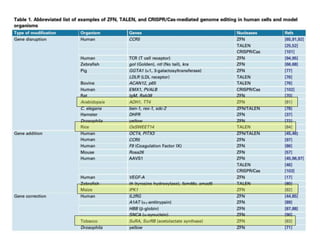
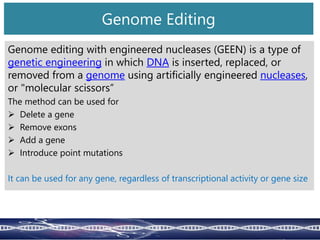
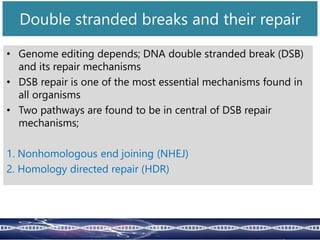
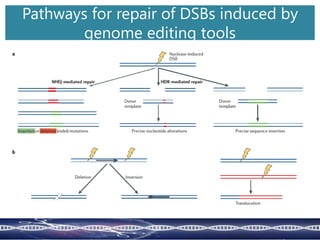
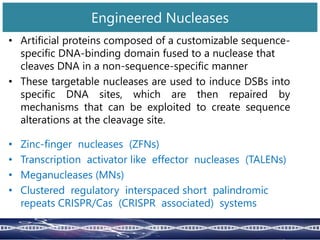
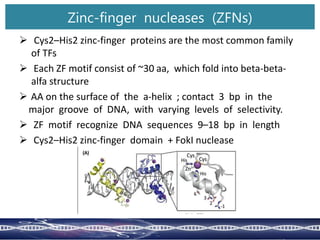
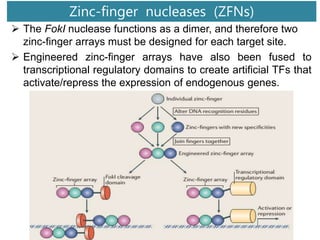
Genome editing uses engineered nucleases to make targeted changes to DNA. There are several methods for engineered nucleases: zinc finger nucleases (ZFNs) use zinc finger proteins fused to FokI nuclease; transcription activator-like effector nucleases (TALENs) use transcription activator-like effector proteins fused to FokI; meganucleases are naturally occurring enzymes; and CRISPR/Cas uses a bacterial adaptive immune system. These nucleases make double-stranded breaks that are repaired through non-homologous end joining or homology-directed repair, allowing changes like gene knockouts, insertions, or replacements. Examples showed using these methods in plants and animals. Each method

![An Example of ZFN technology
ZFNs used to cleave and stimulate mutations at an endogenous target gene [ABA-
INSENSITIVE4 (ABI4)] in Arabidopsis
This gene controls a number of agronomically important traits, including plant responses
to abiotic stress and seed development
They achieved targeted mutagenesis at a rate of ~ 0.26% to 2.86% in Arabidopsis somatic
cells, and transmission of the induced mutation in the target gene to subsequent
generations
Consensus ZFN target sites in the Arabidopsis ABI4
gene. Asterisk indicates the position of the mutation
in the abi4 mutant. Target sites of ZFN monomers
are highlighted with gray bars. The putative
cleavage sites are shown by arrows.](https://image.slidesharecdn.com/genomeeditingpresentation-230921054845-027103b4/85/Genome-editing-presentation-pptx-9-320.jpg)