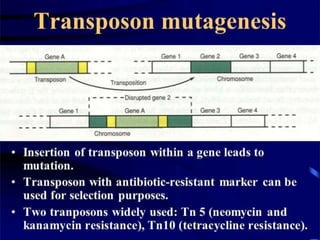
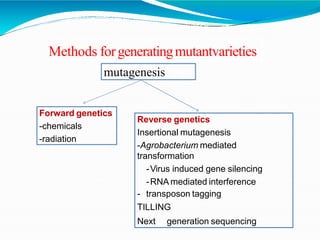
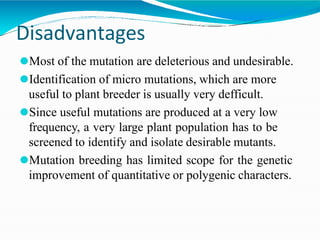
The document provides a comprehensive overview of mutagenesis, detailing its types, applications, and methods for screening and validation of mutants. It covers various techniques such as site-directed mutagenesis, insertional mutagenesis, and the use of genetic screens for studying gene functions. Additionally, it discusses both the advantages and limitations of mutation breeding as well as future prospects for using mutants in genomic research.