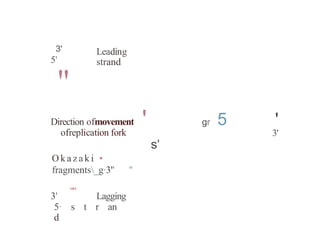
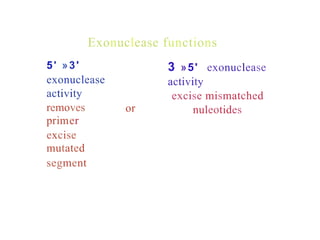
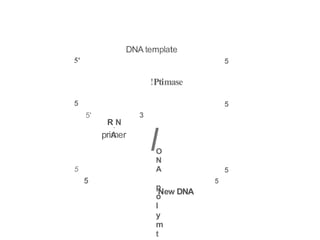
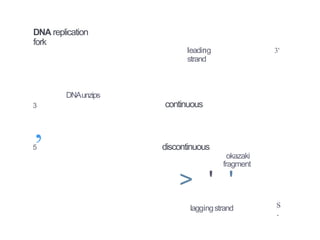
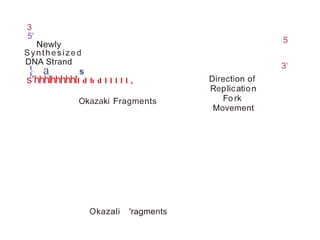
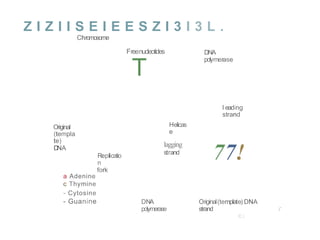
The document describes the central dogma of molecular biology, focusing on DNA replication, which is a crucial process for biological inheritance. It outlines the mechanisms of semiconservative replication, including the roles of various enzymes such as DNA polymerase, helicase, and primase, as well as details on replication patterns and fidelity. Finally, it explains the steps of initiation, elongation, and termination during DNA replication.







![[1958]
demonstrated Semiconservative
replication
DNA o rtreted an d cen t ri fu g ed
to u t i t r u r n n € C T d n a t t y
g r a d i n t
o
r
i
g
i
n
8a
lp a r e n t
t
n o . c u l
Mytr@ DNA
M N I4N»
ct»
t o r t i - o n
d u g h t r r o e l o
3t
l
c
o
#nd
e
3t
e
n
8t
i
o
n
L e
D N A ( ' N
t e H y b r i d D N A](https://image.slidesharecdn.com/dna-replication-241016180523-37aac501/85/DNA-synthesis-molec-DNA-replication-pptx-9-320.jpg)