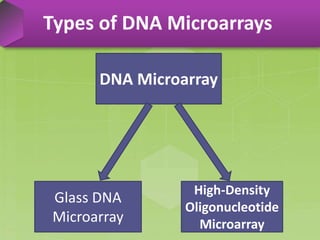
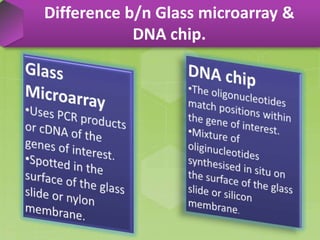
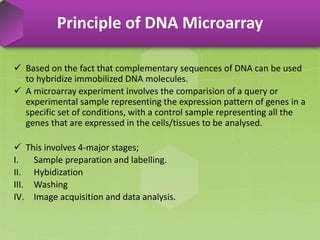
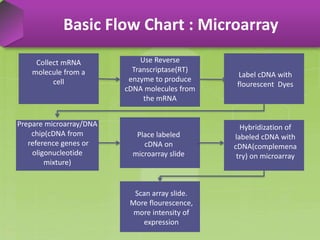
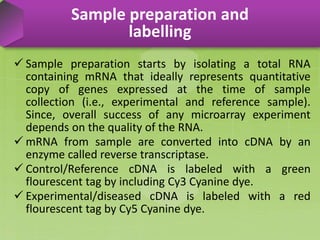
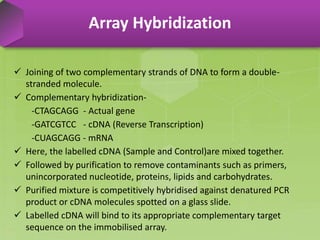
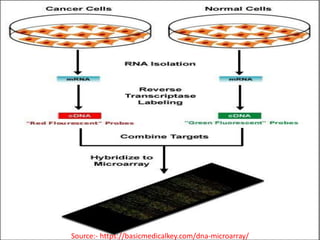
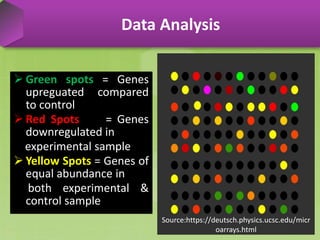
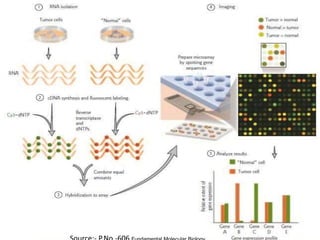
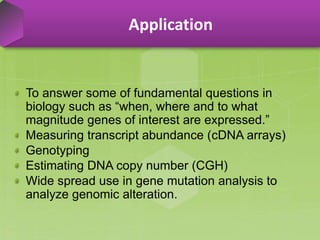
DNA microarrays, also known as DNA chips, are solid supports arranged with thousands of sequenced genes and enable the comparative analysis of gene expression. Patrick Brown and colleagues debuted this technology at Stanford University in 1995, paving the way for advancements in molecular biology. The process involves sample preparation, hybridization of labeled cDNA to a microarray, followed by data analysis to observe gene expression patterns.