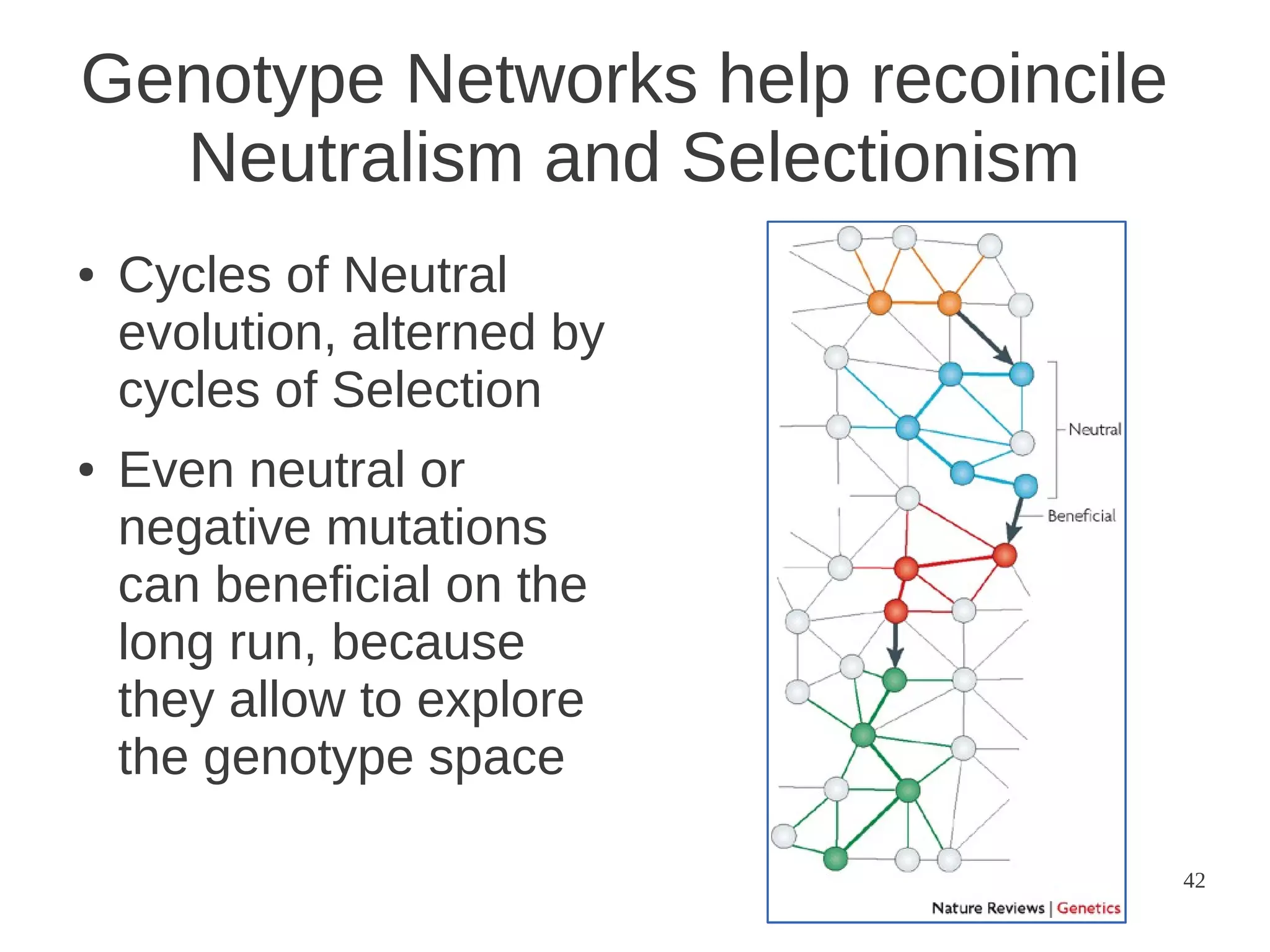
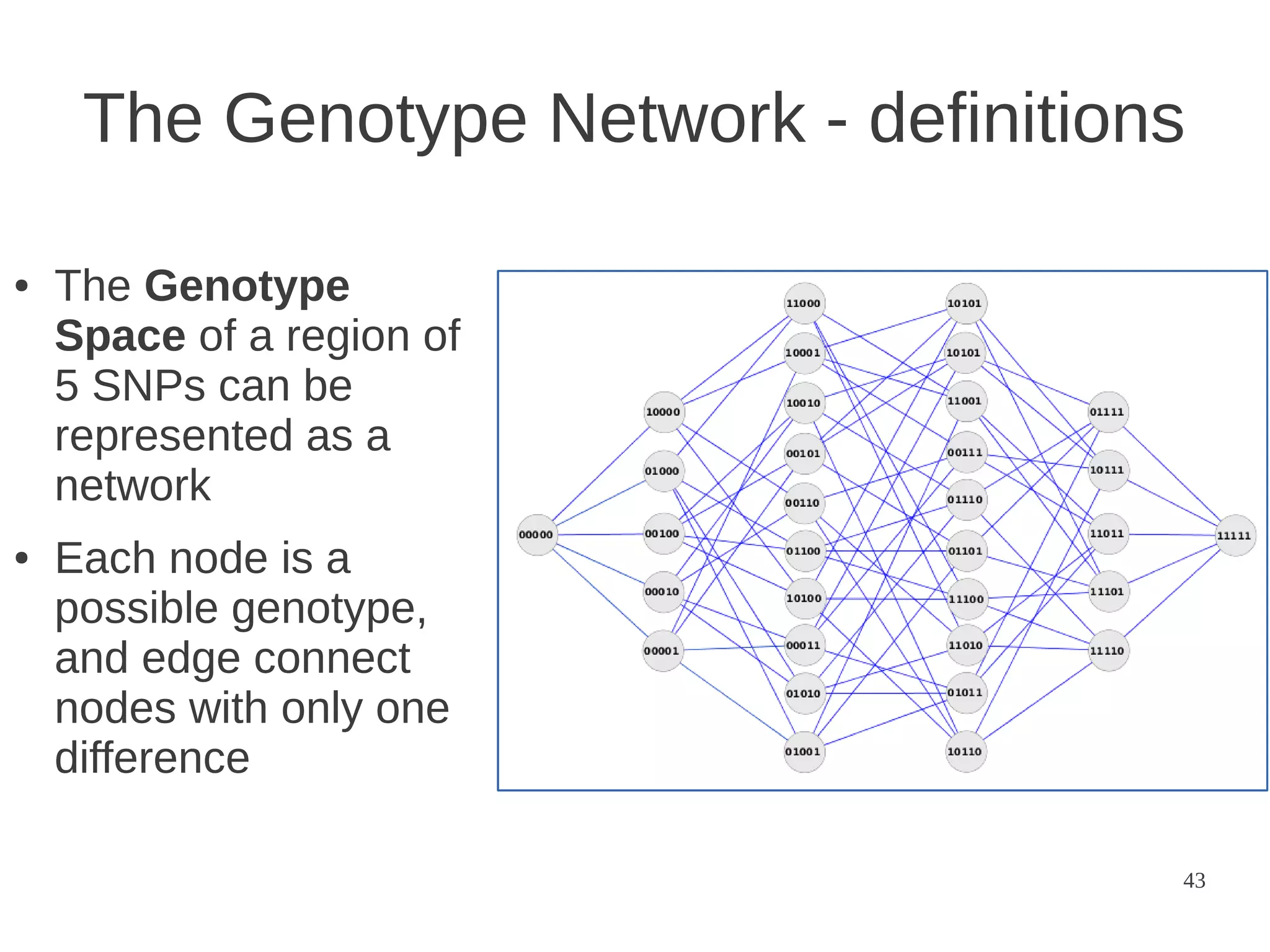
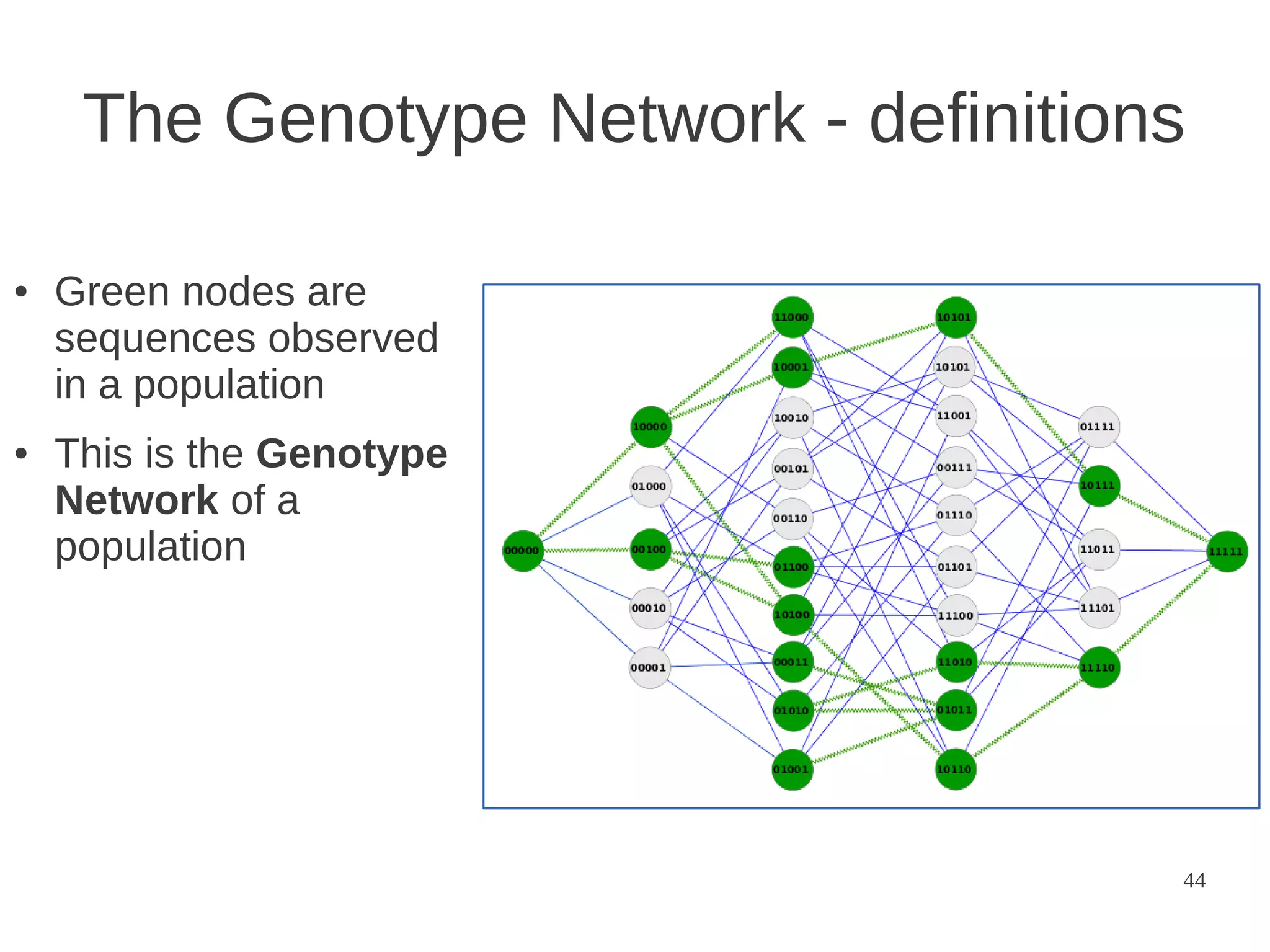
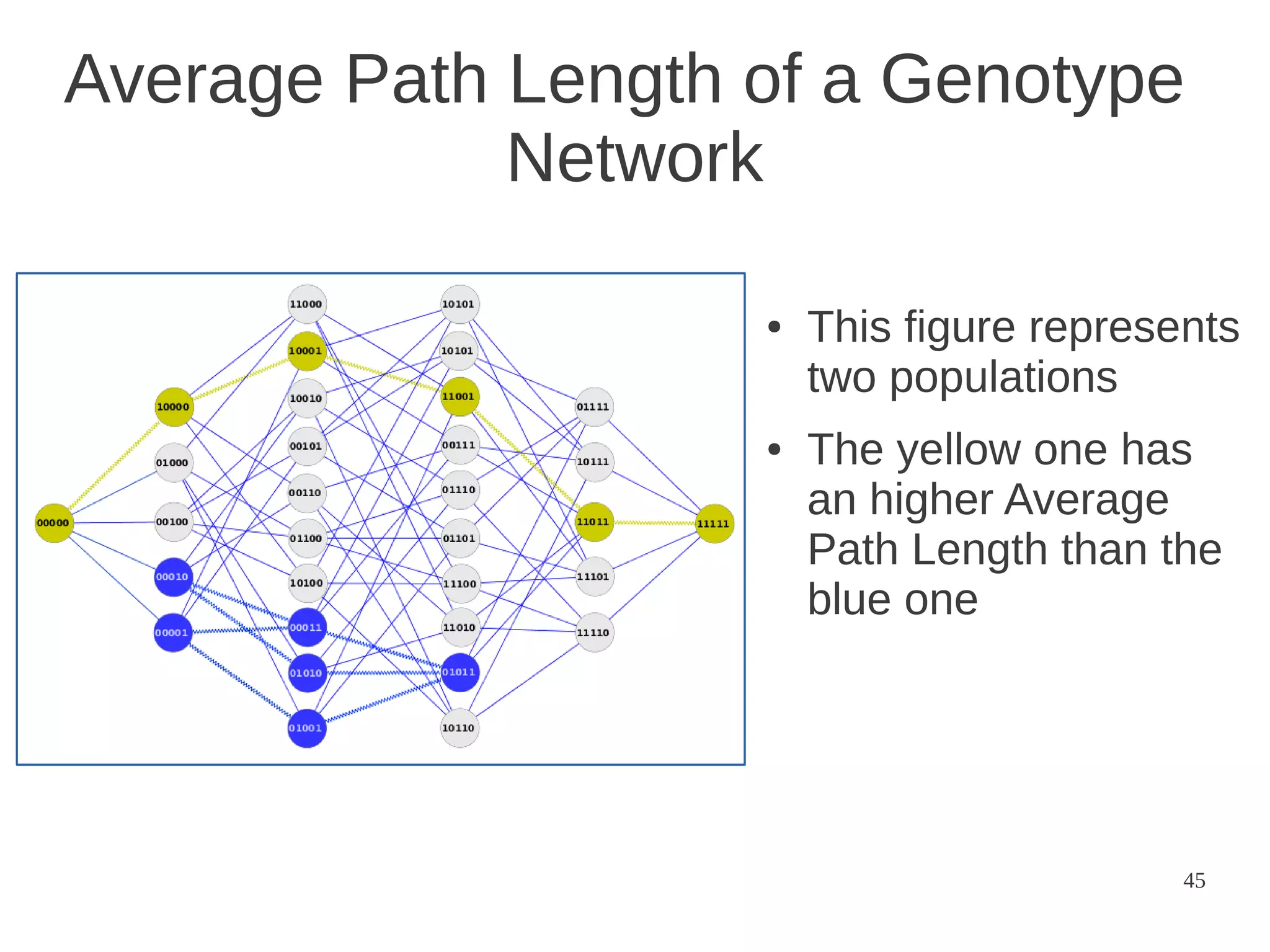
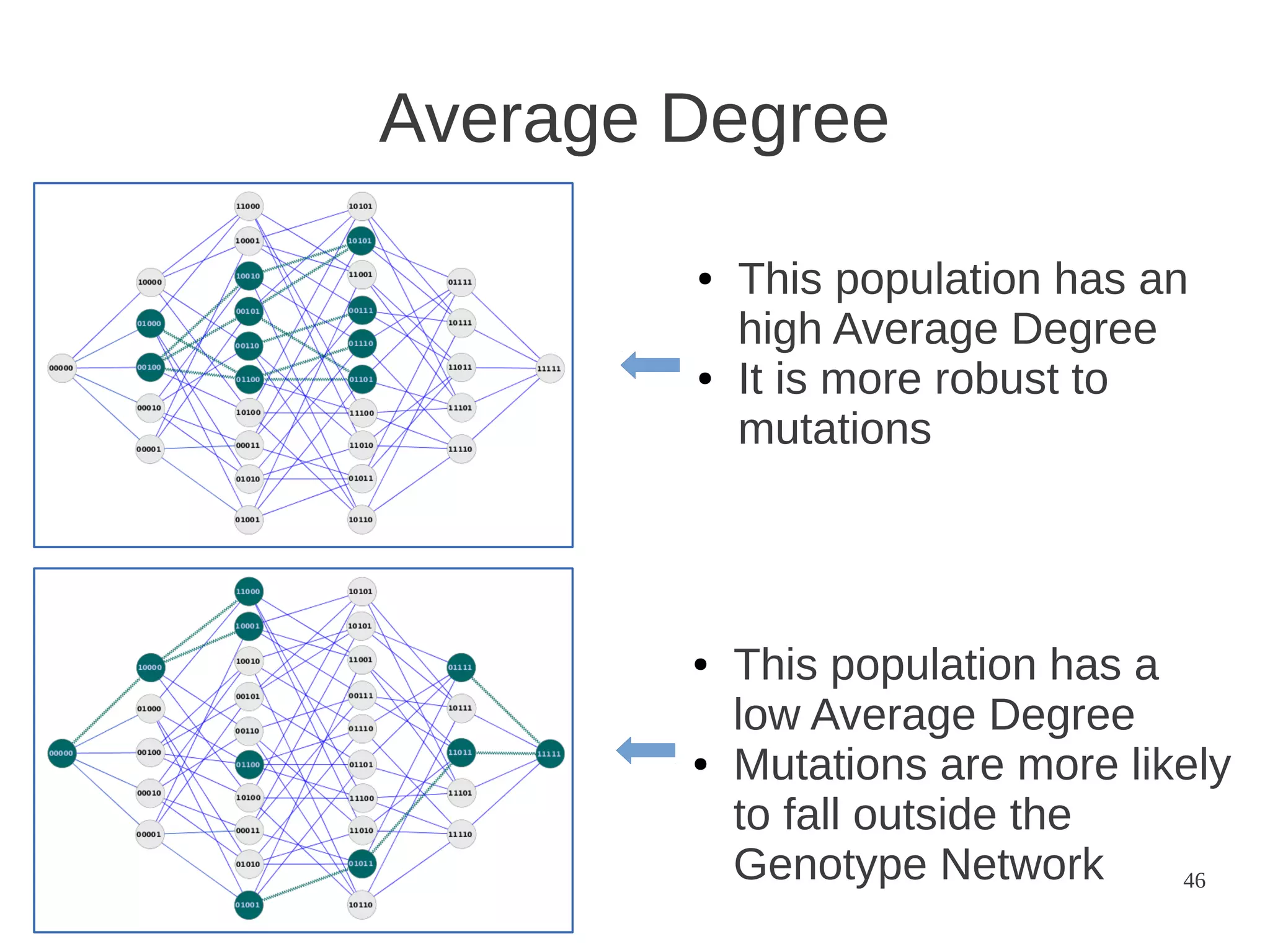
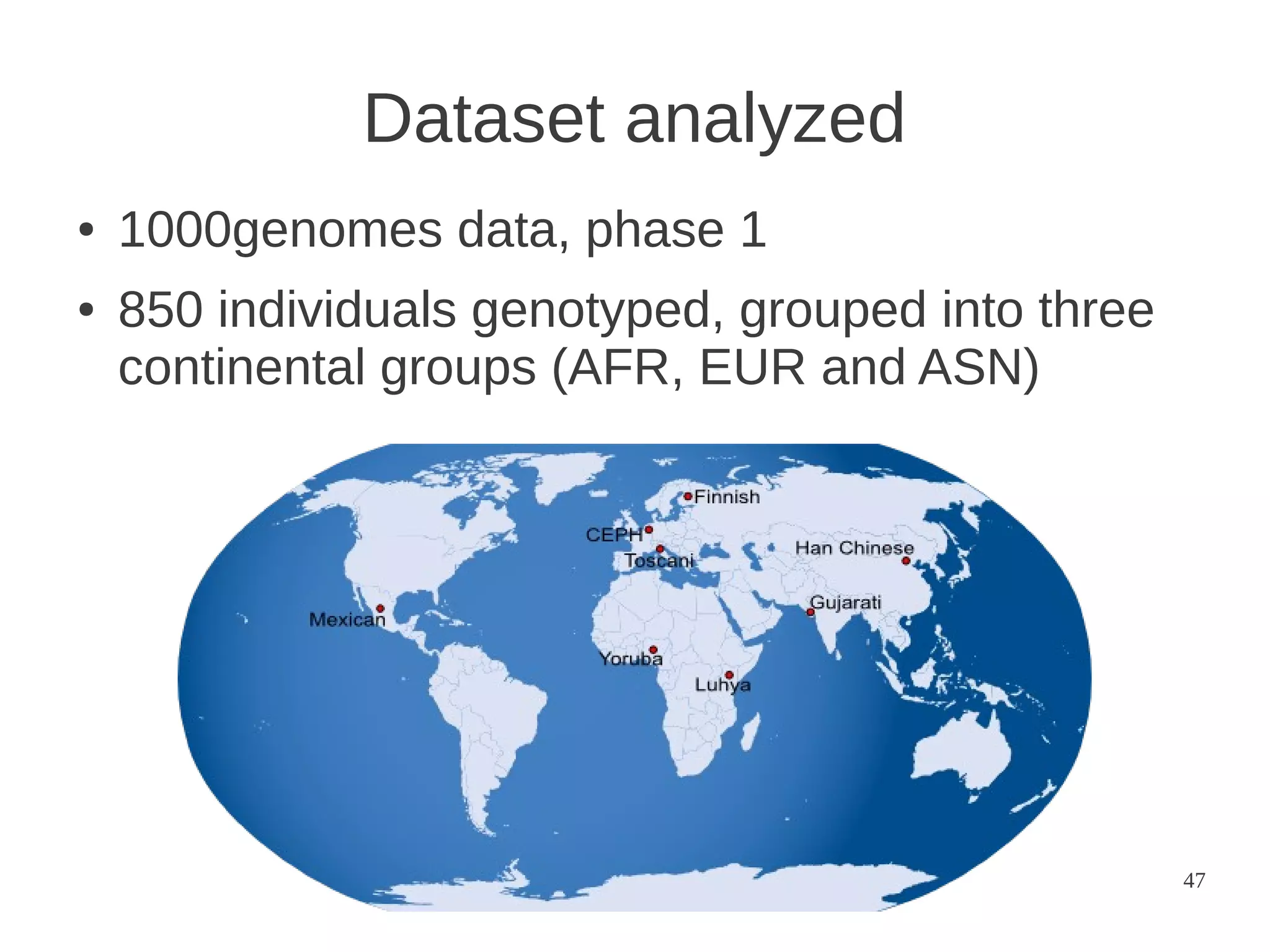
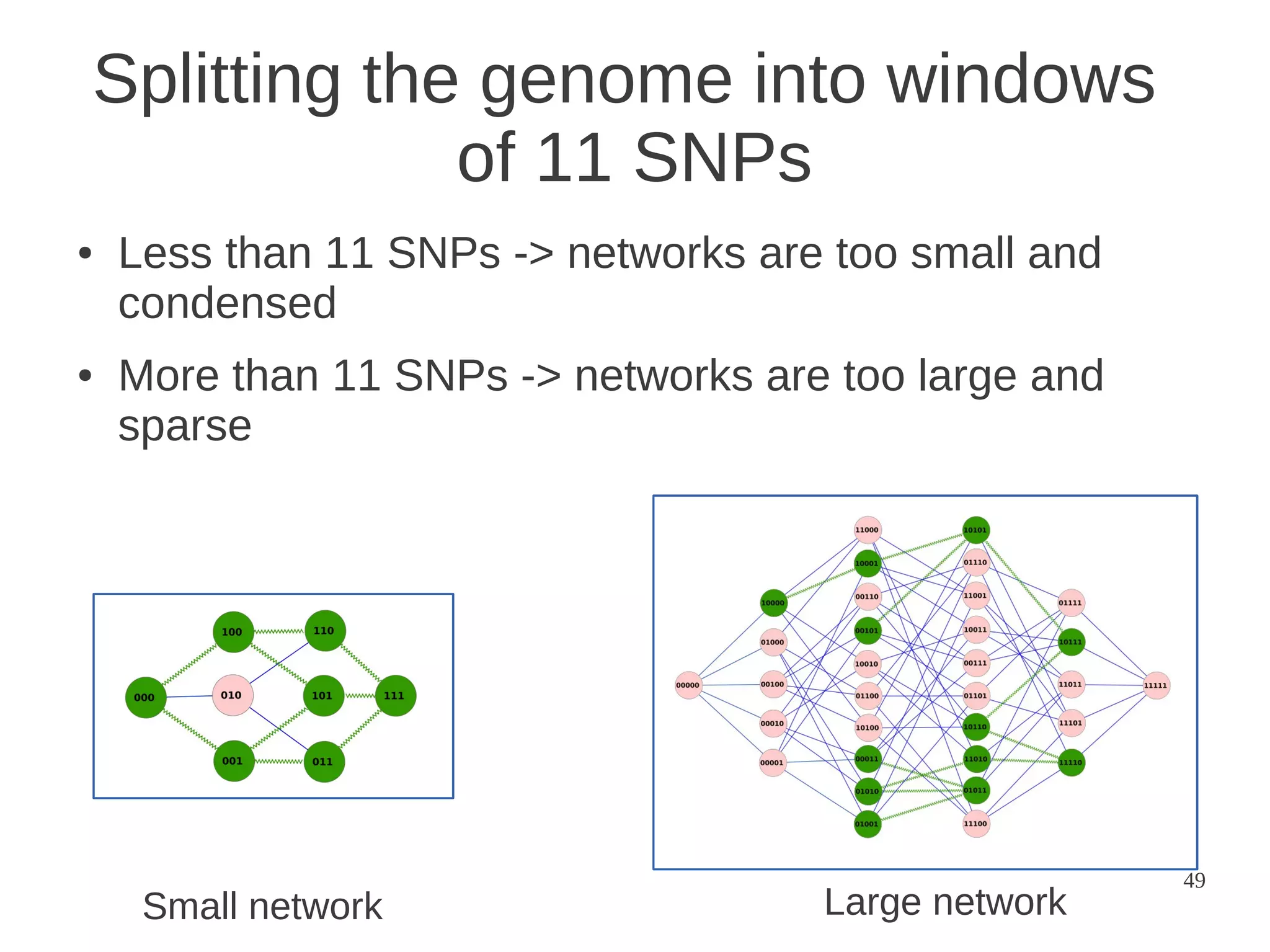
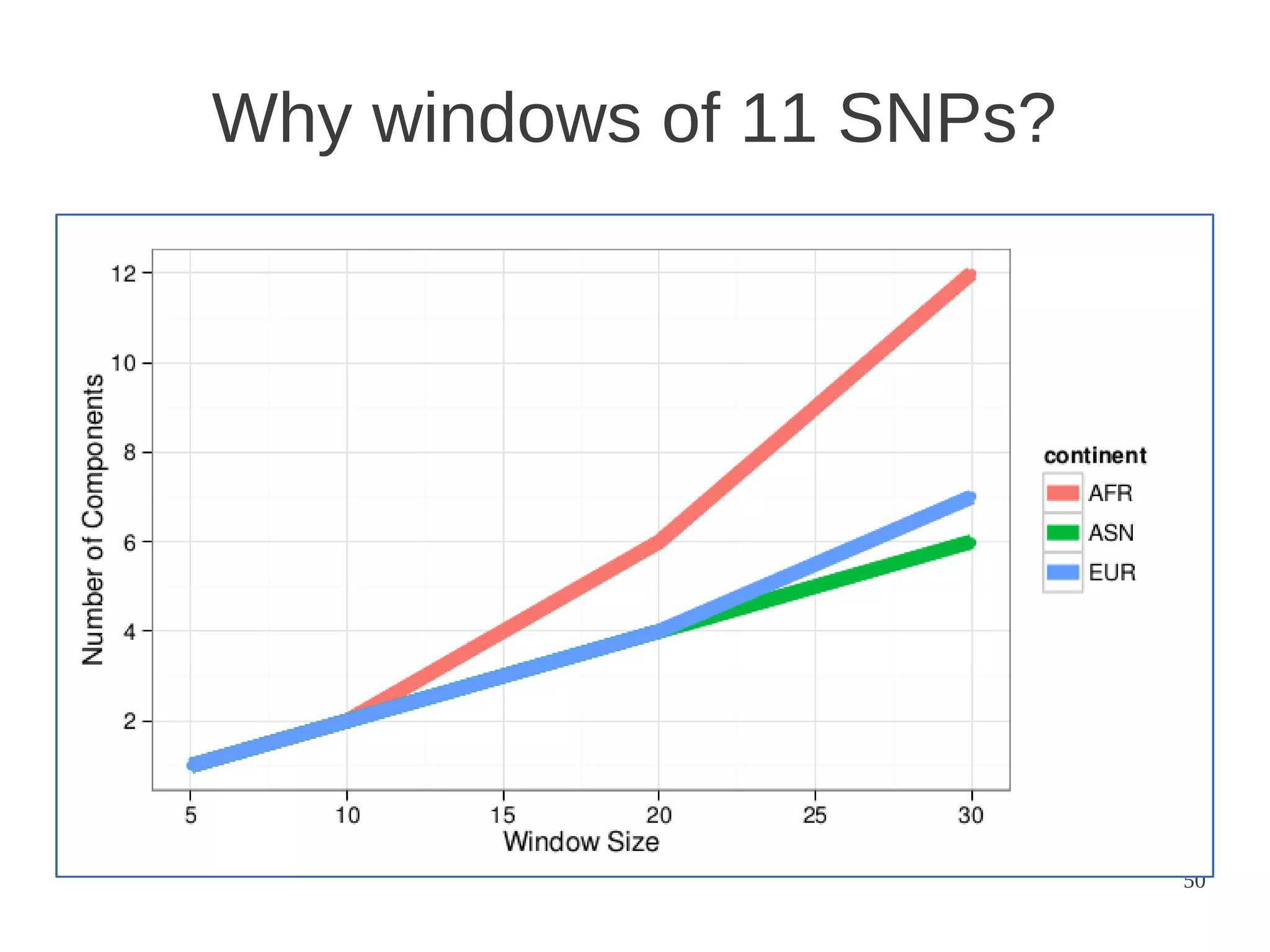
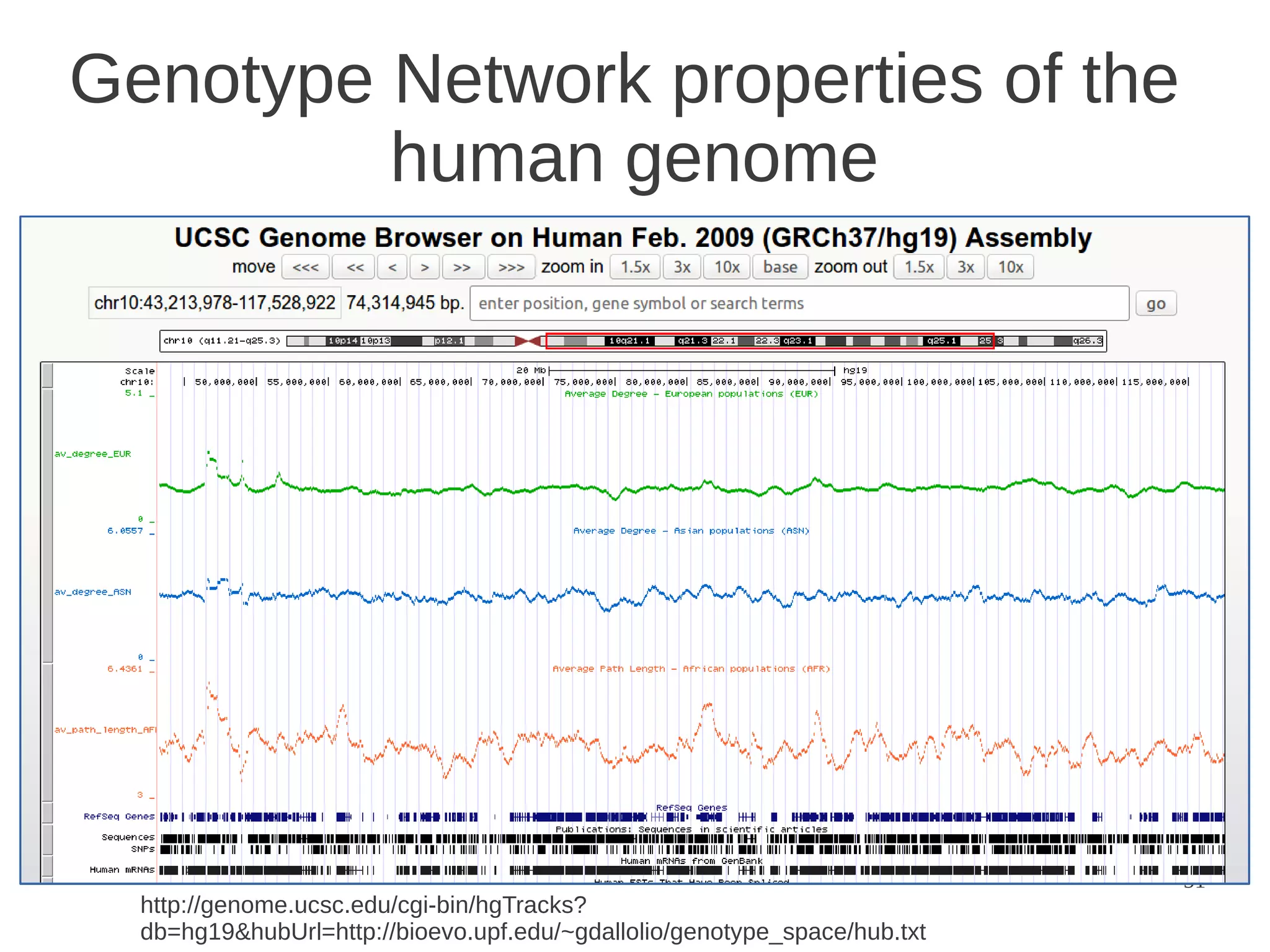
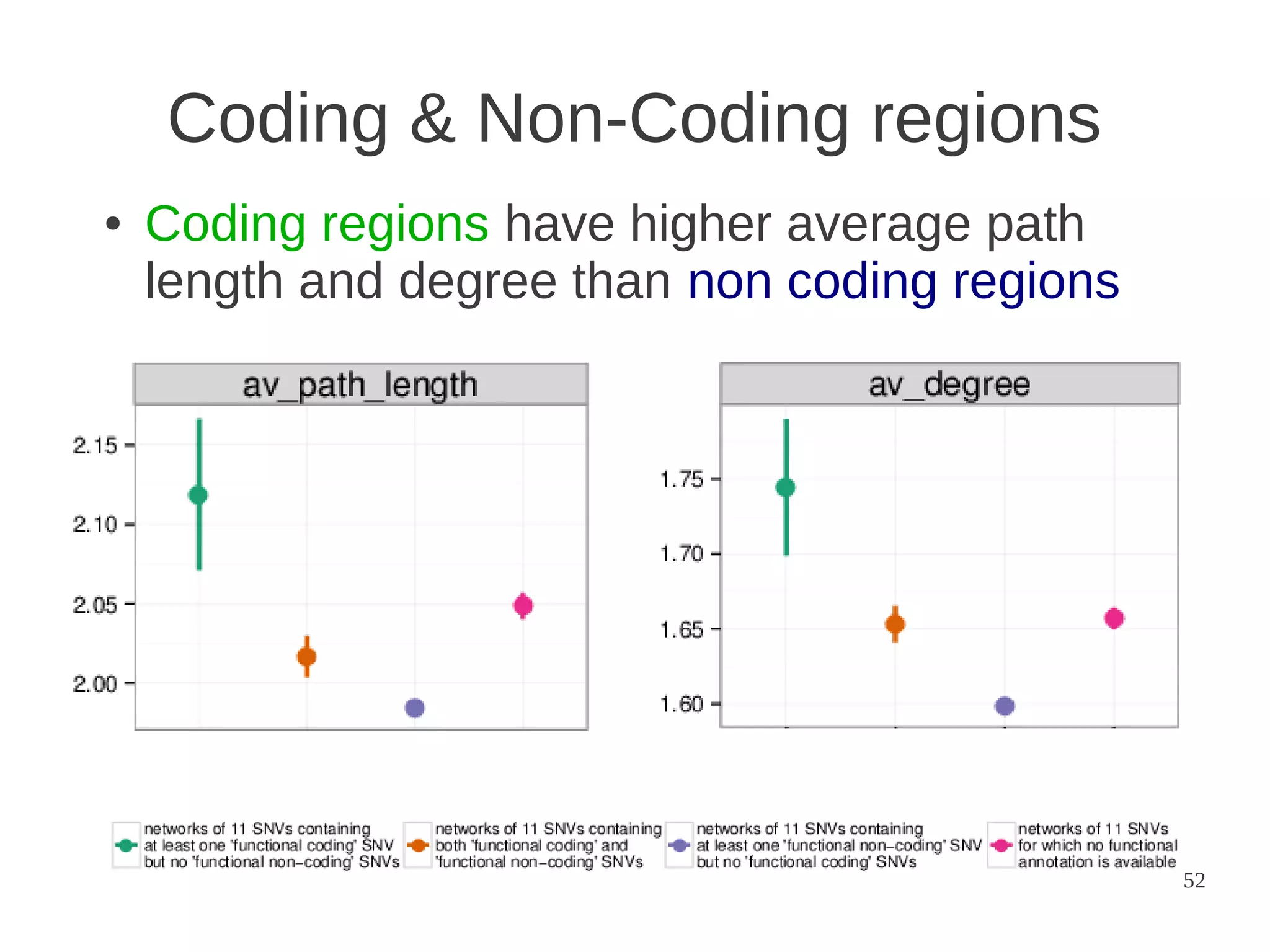
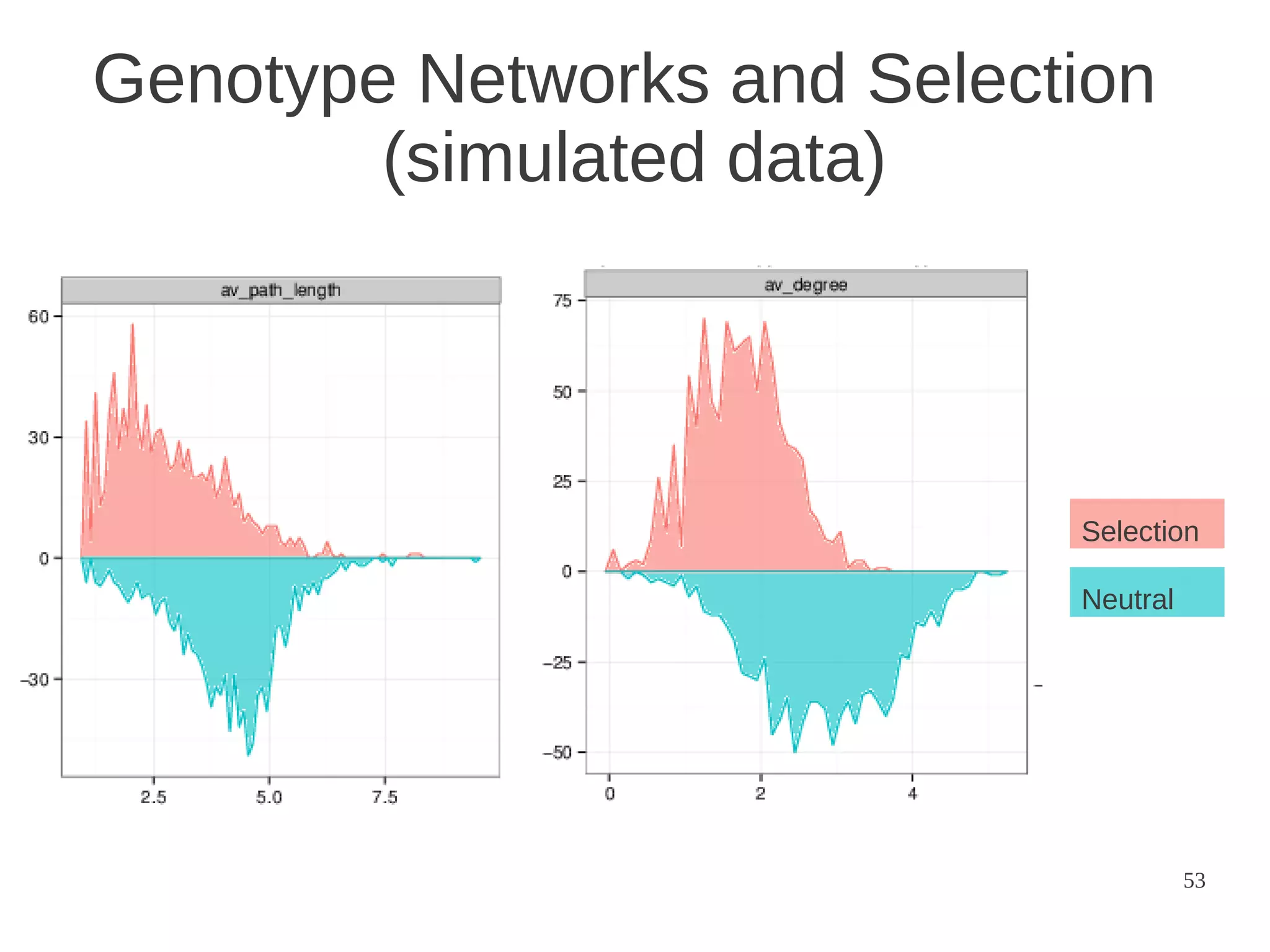
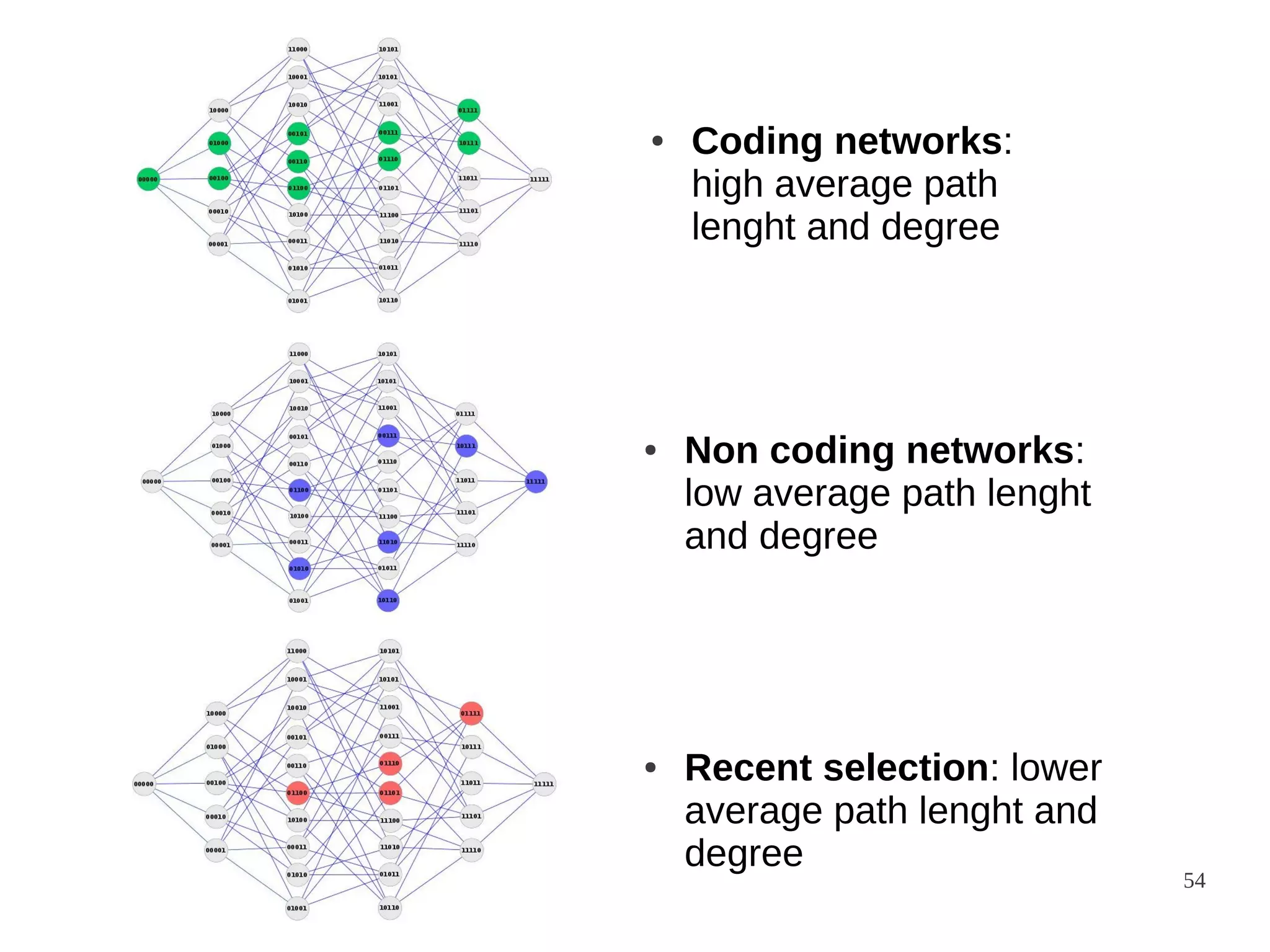
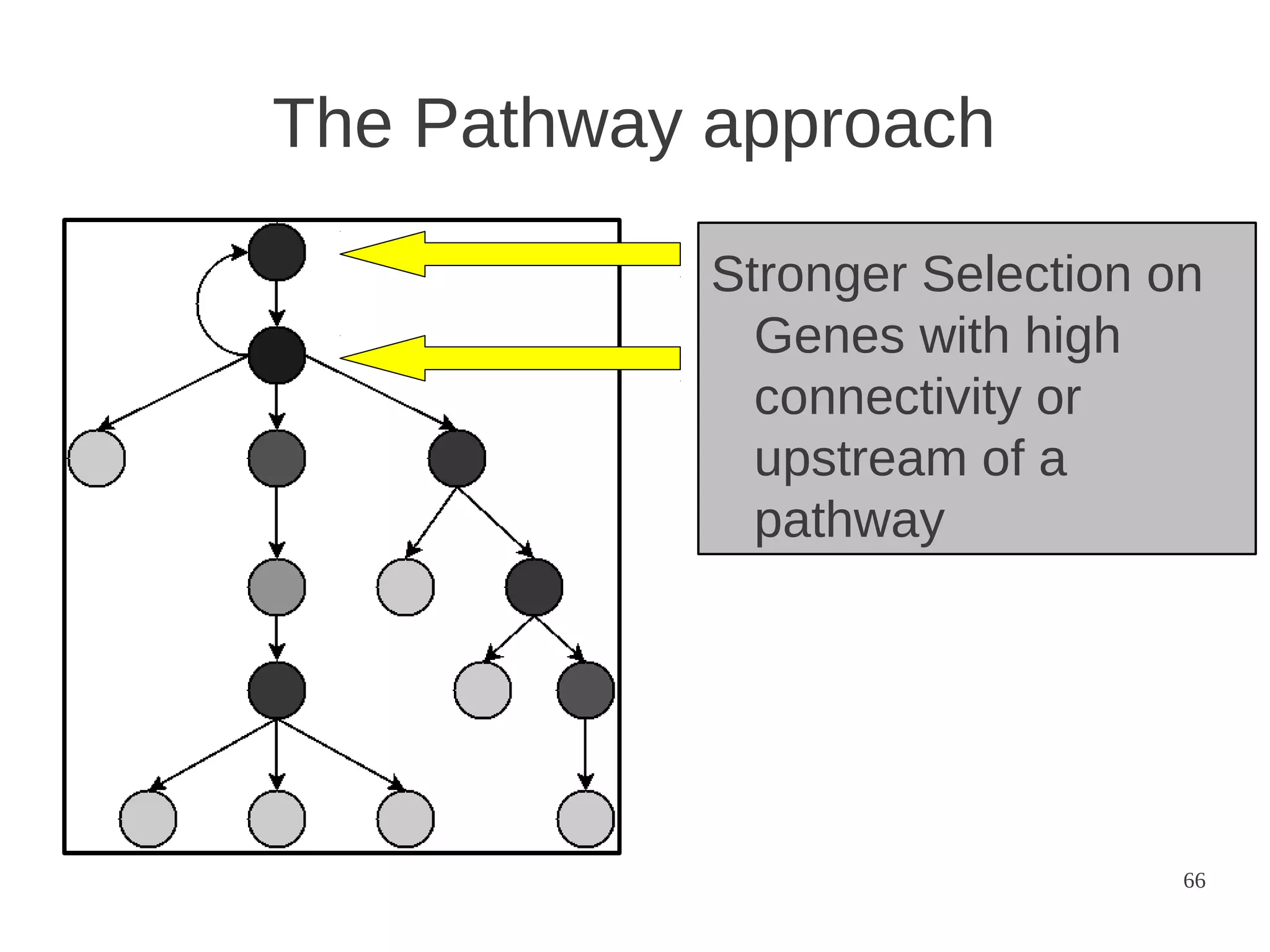
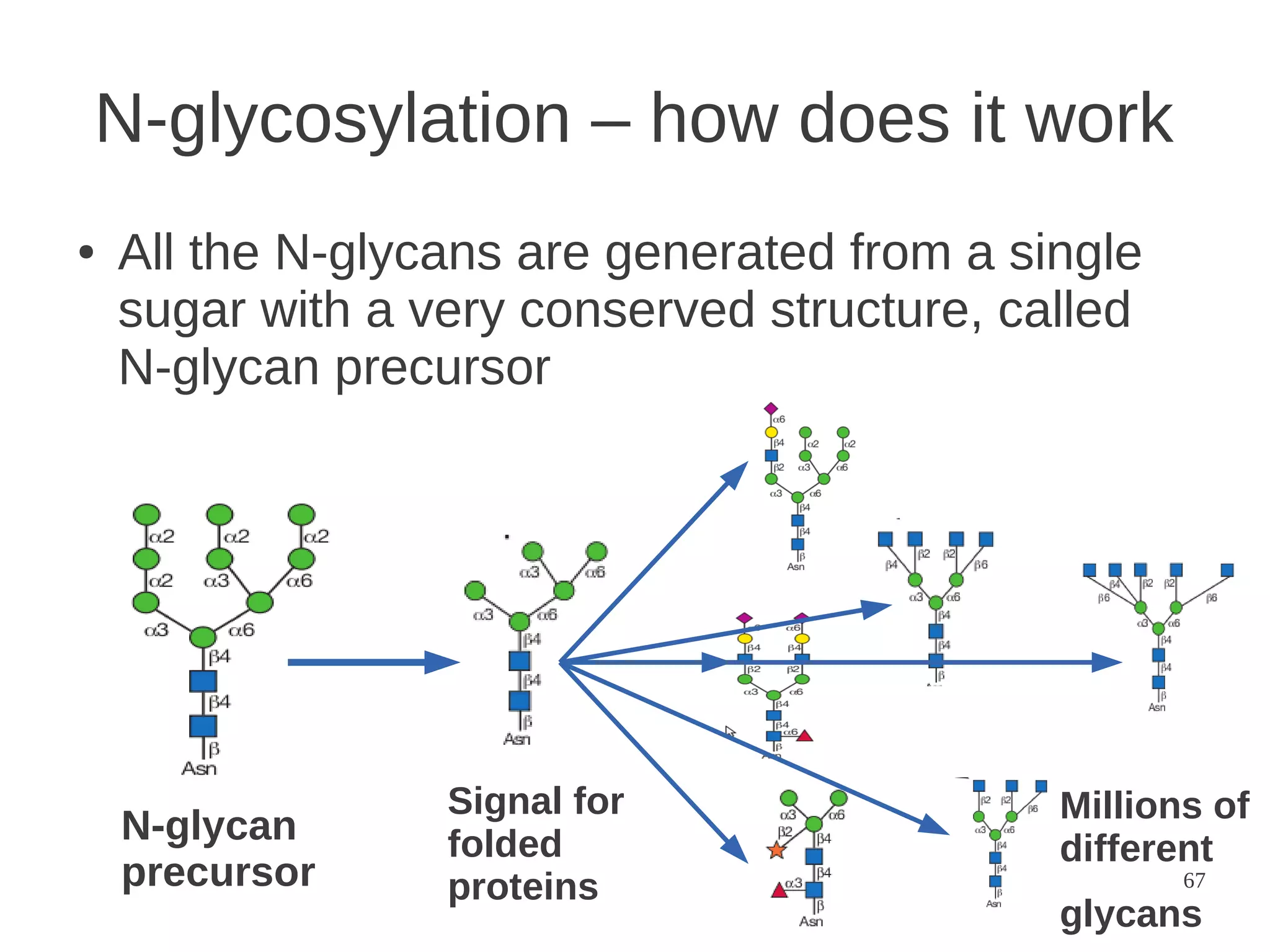
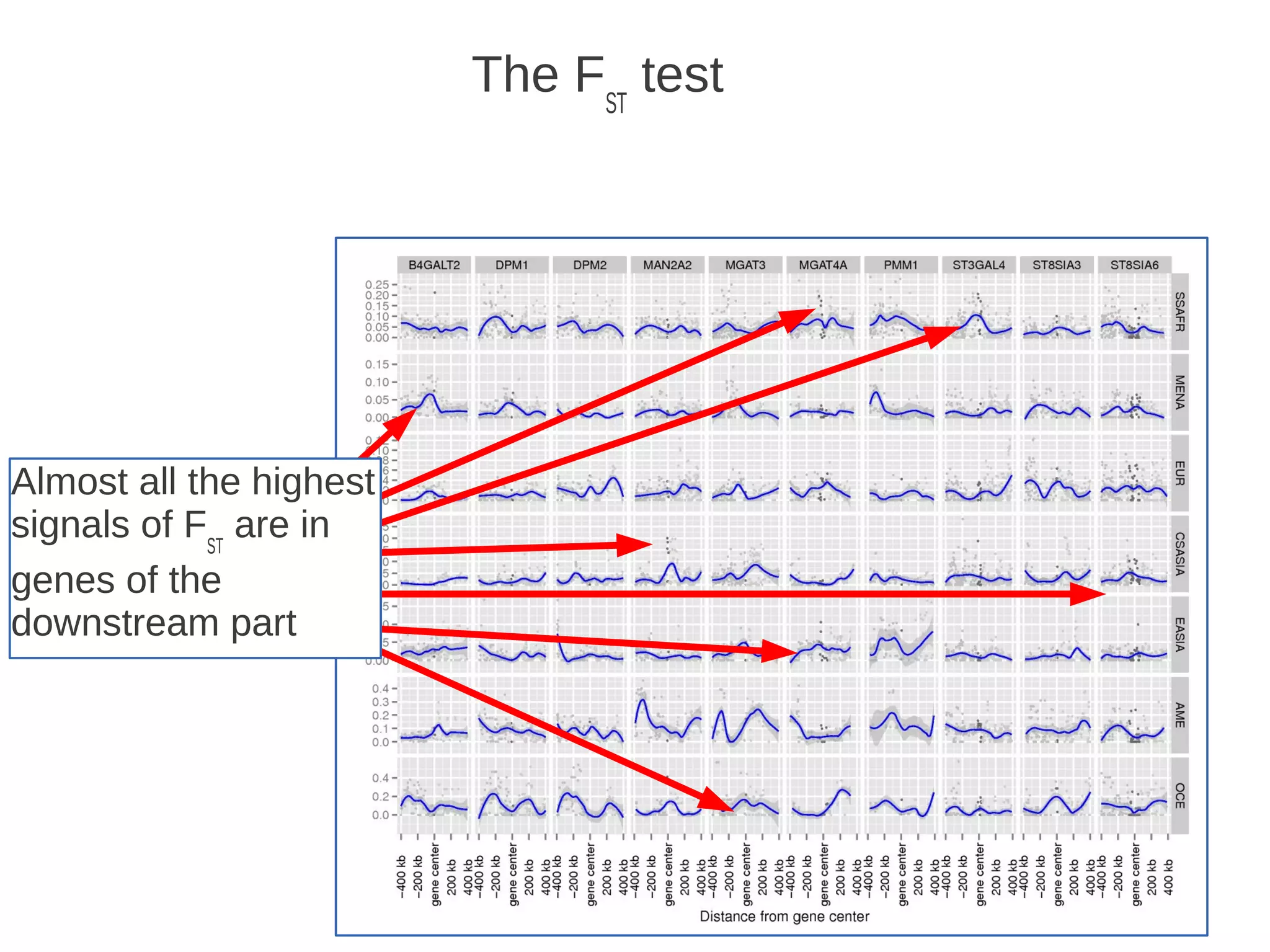
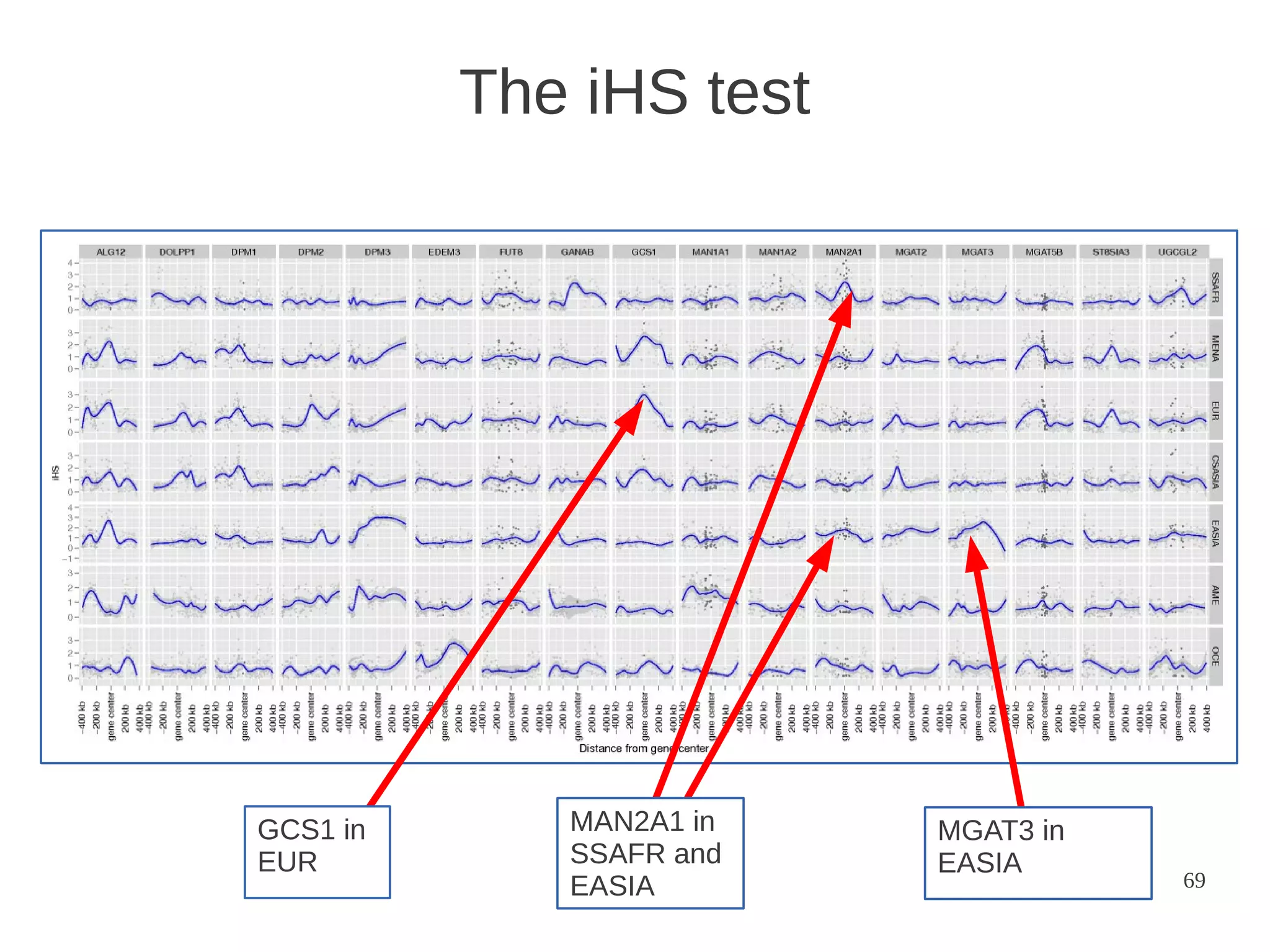
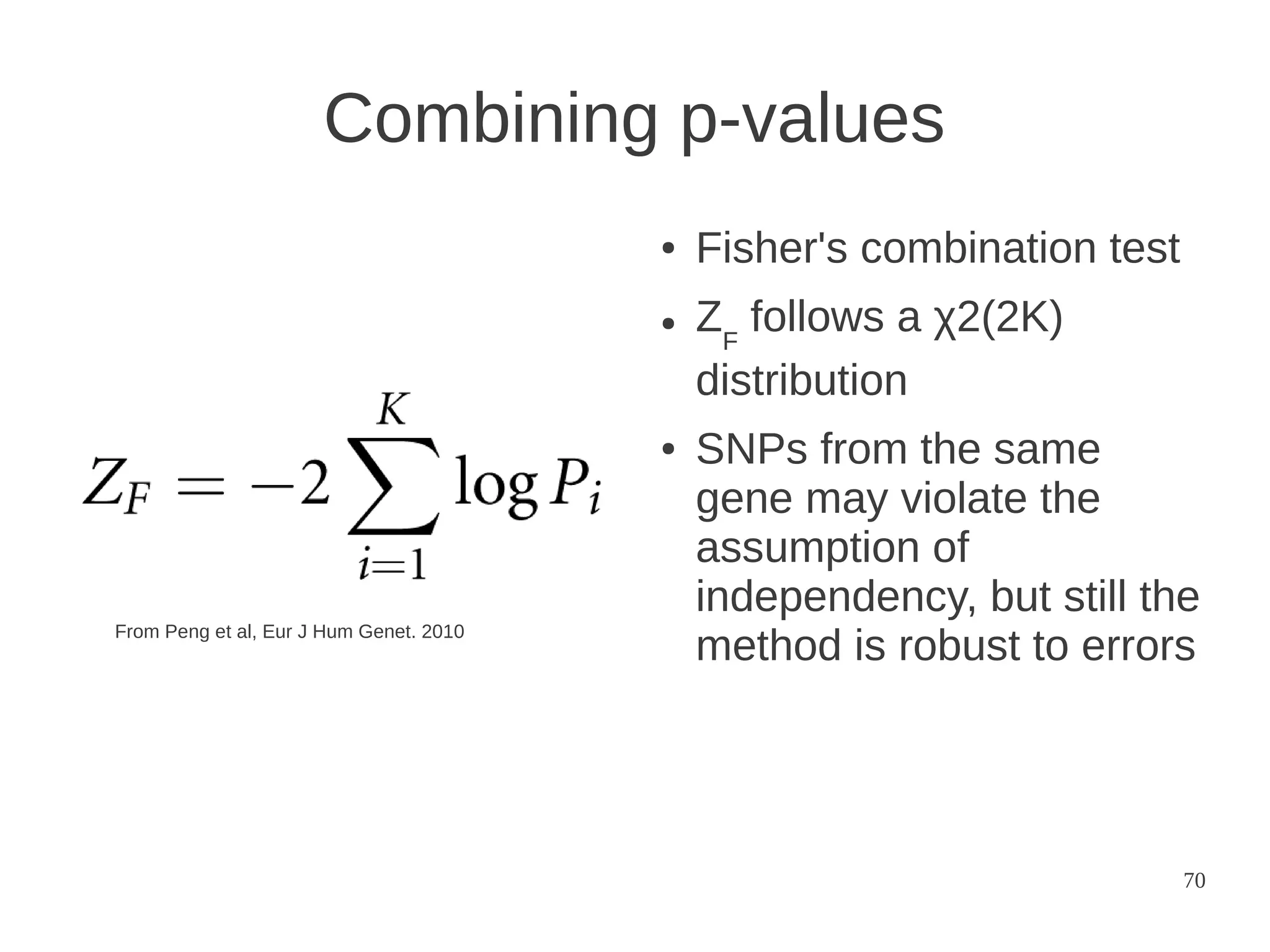
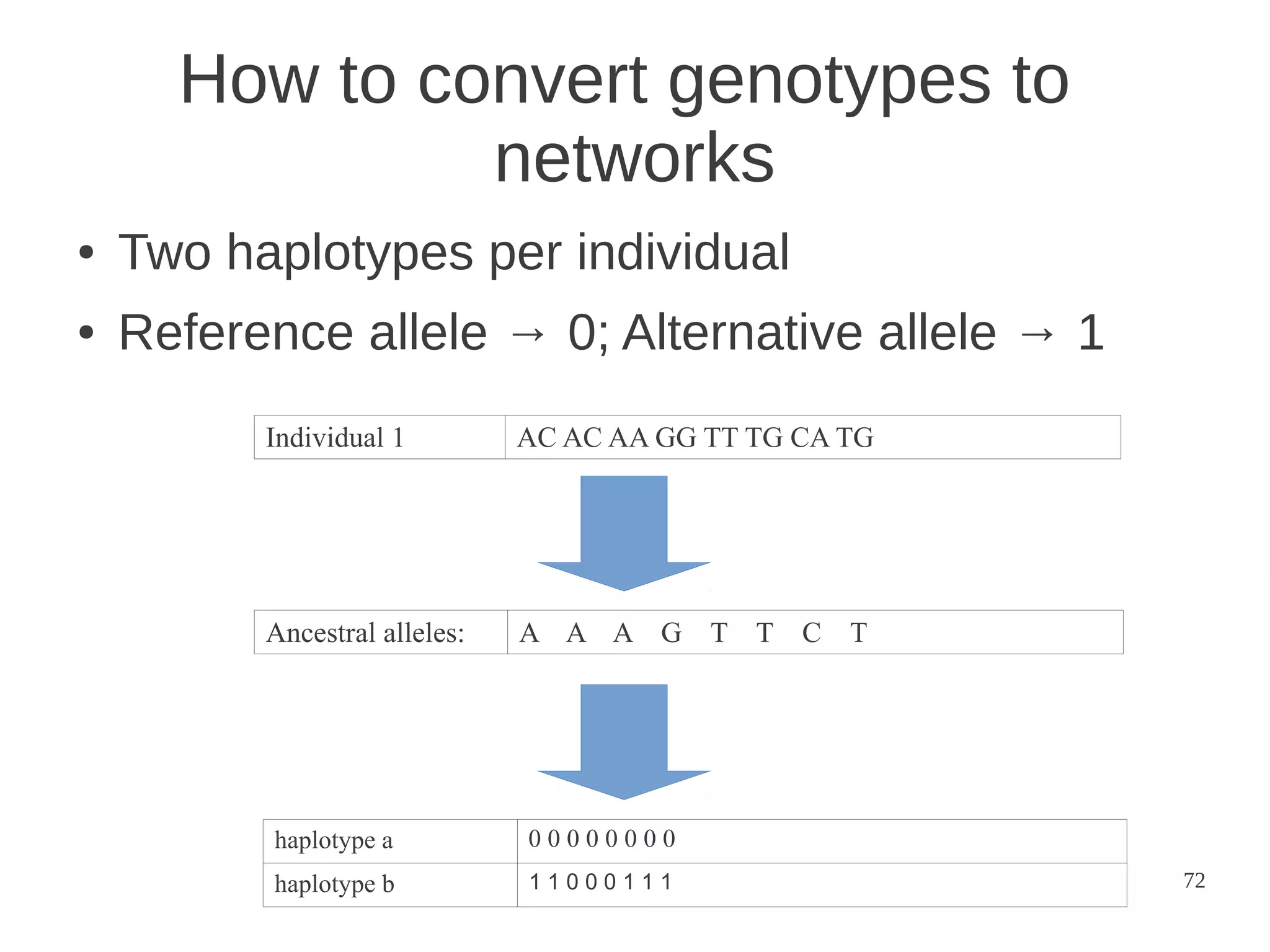
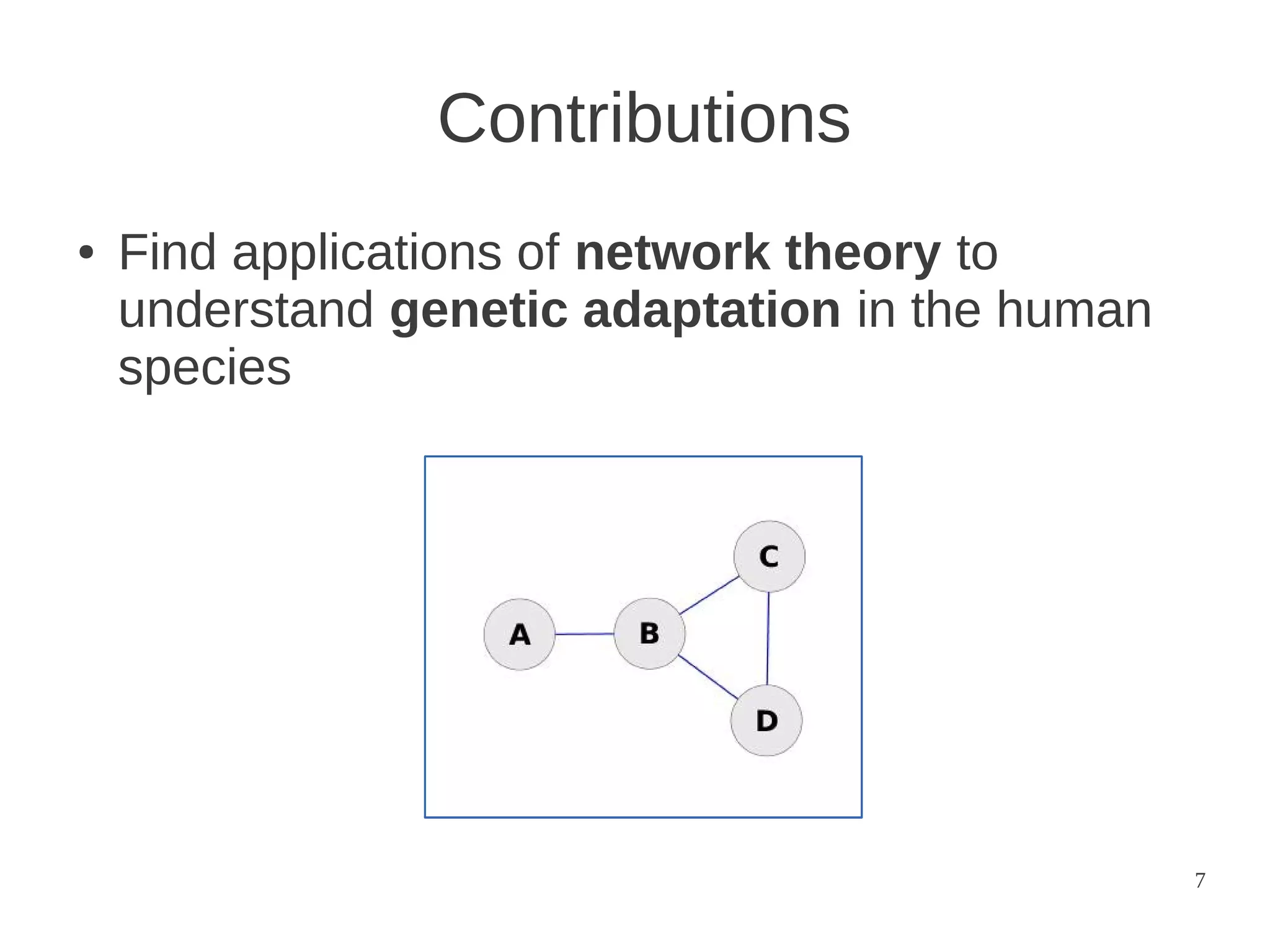
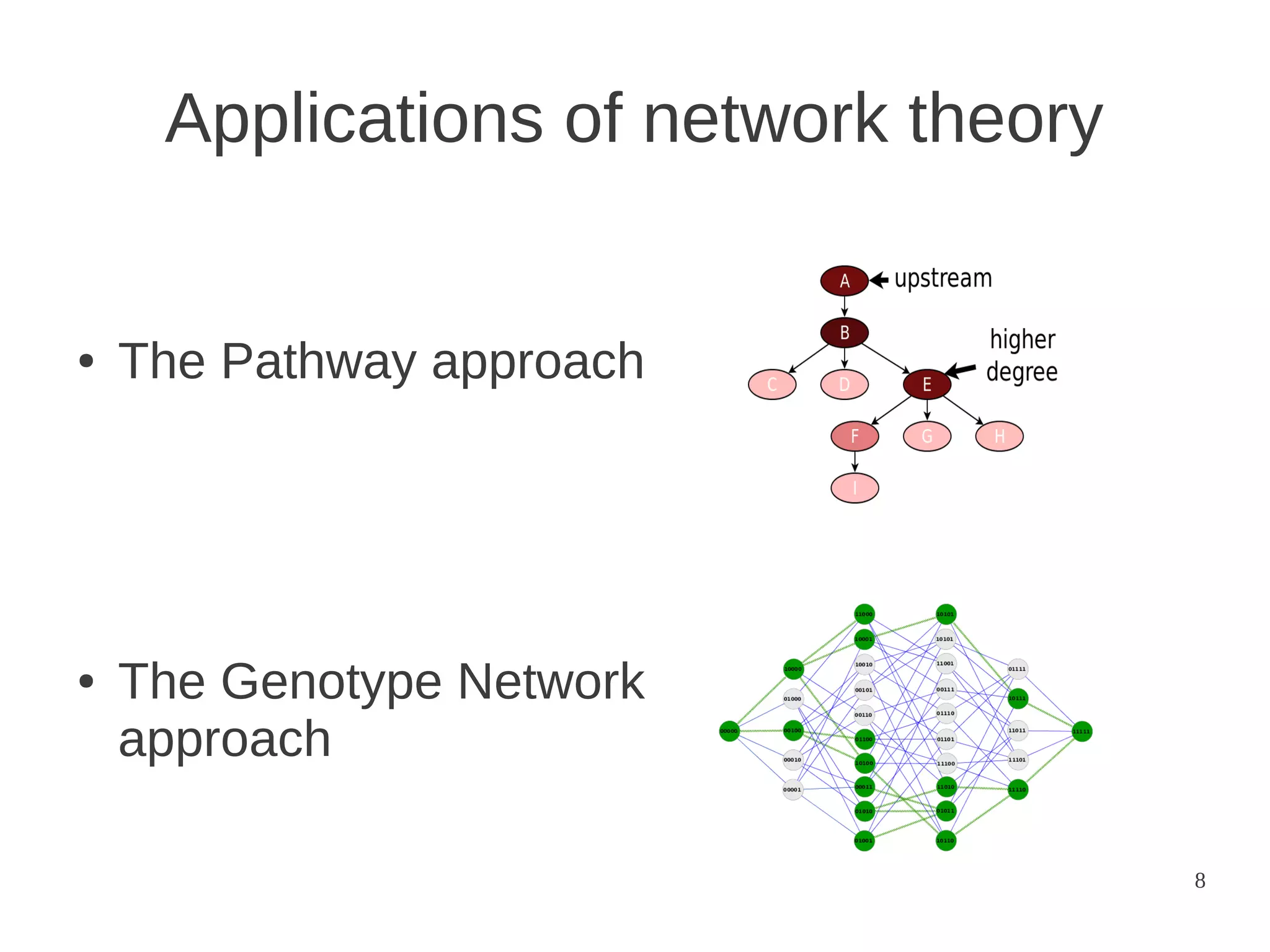
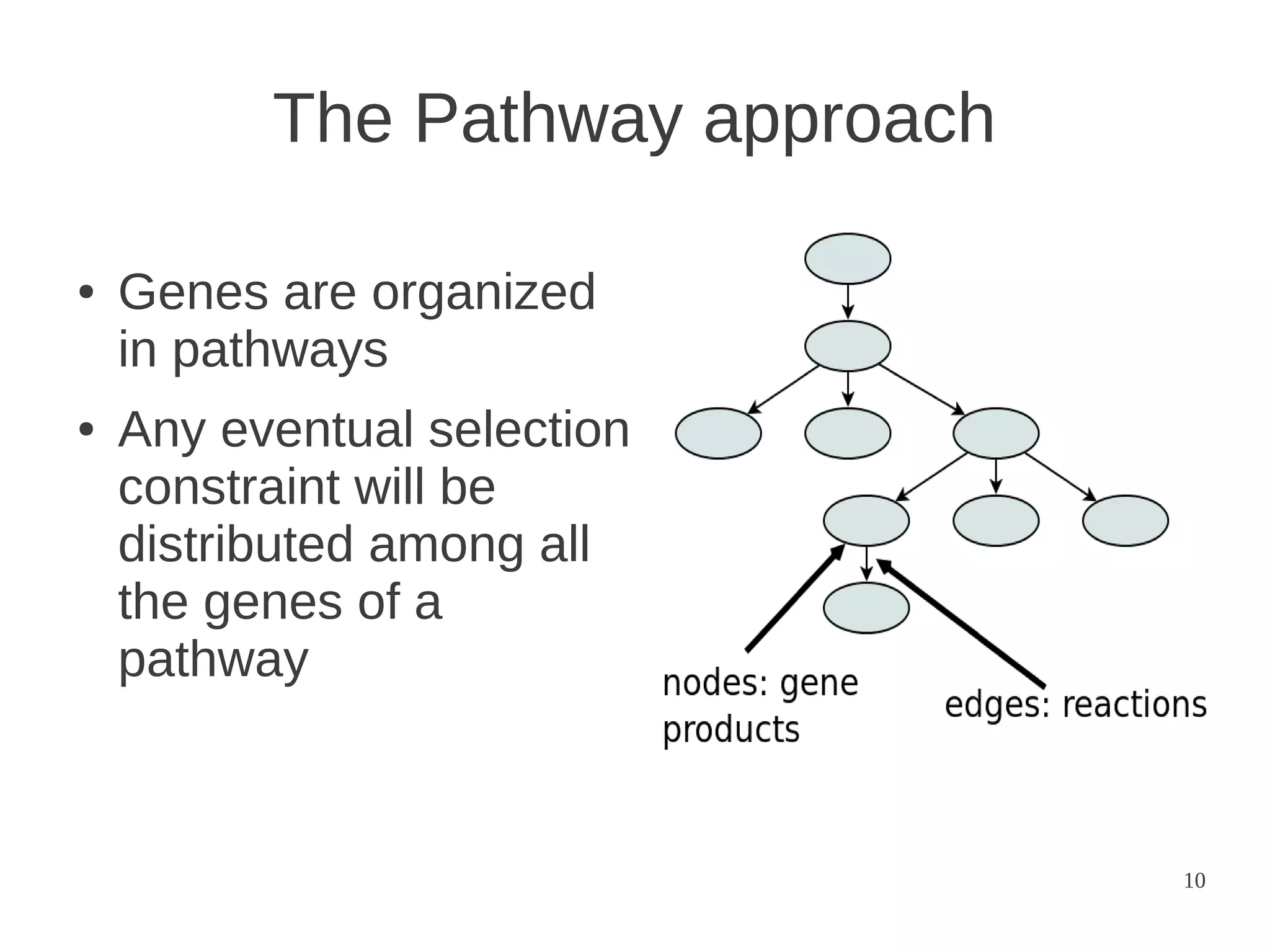
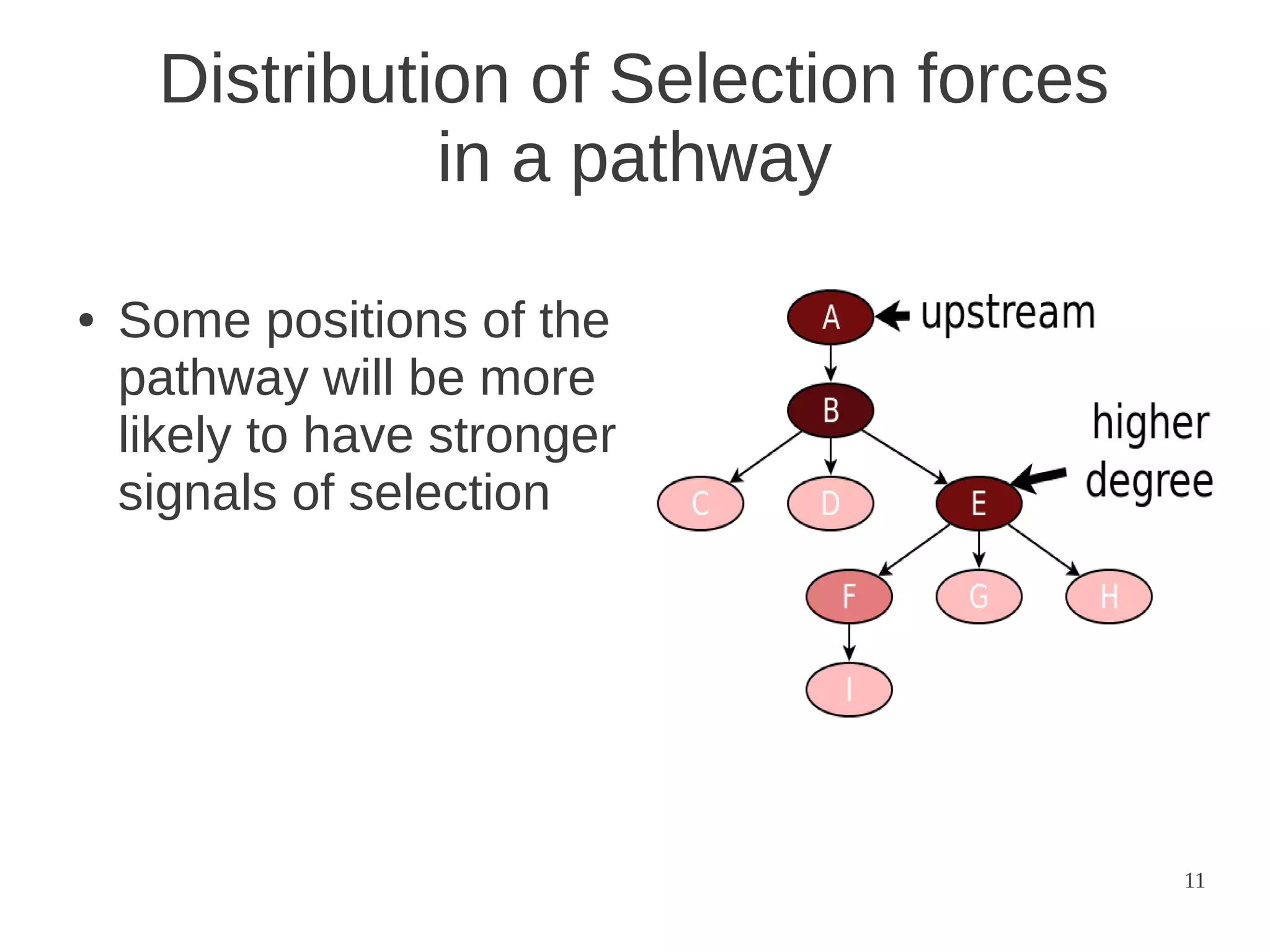
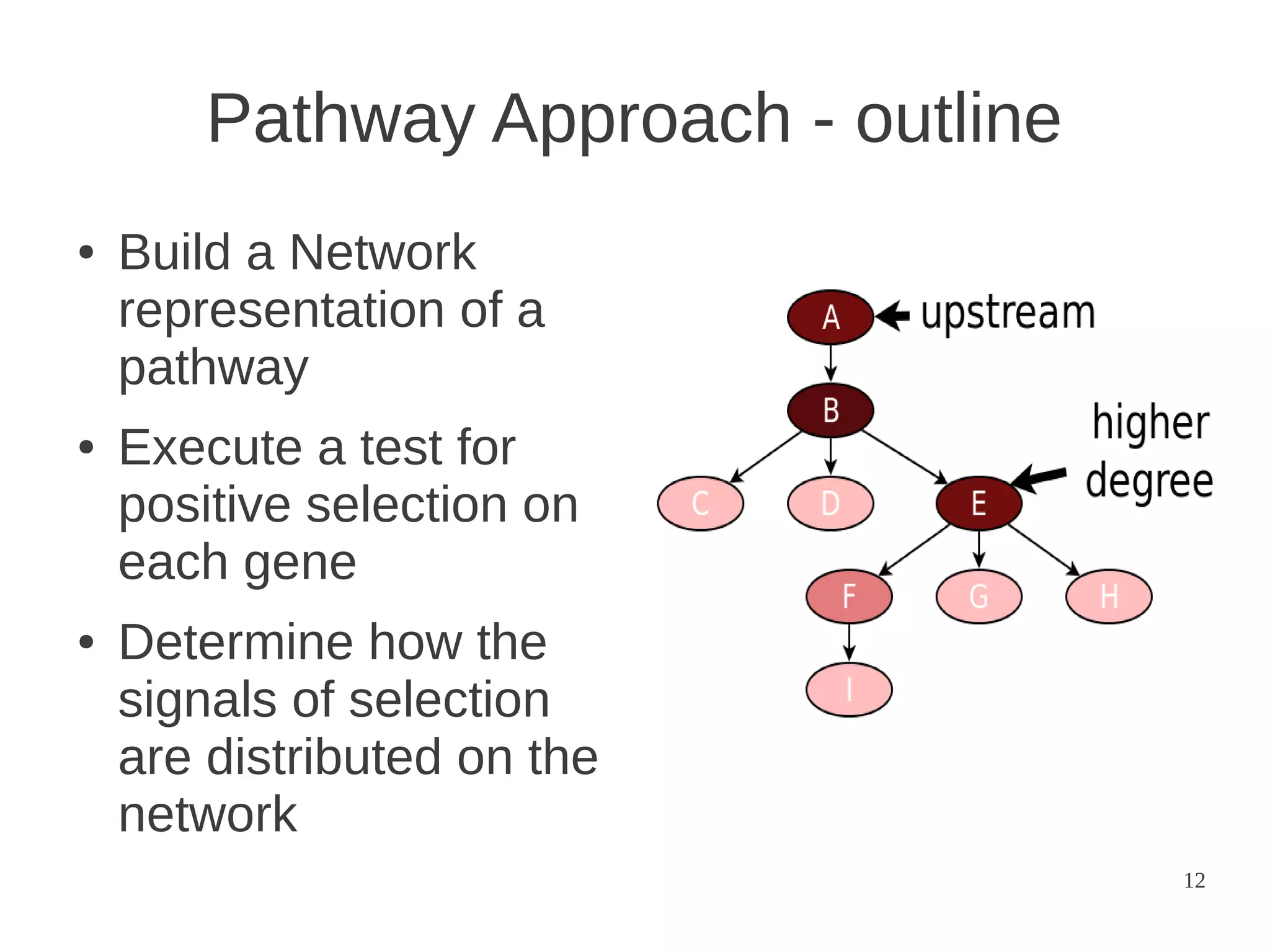
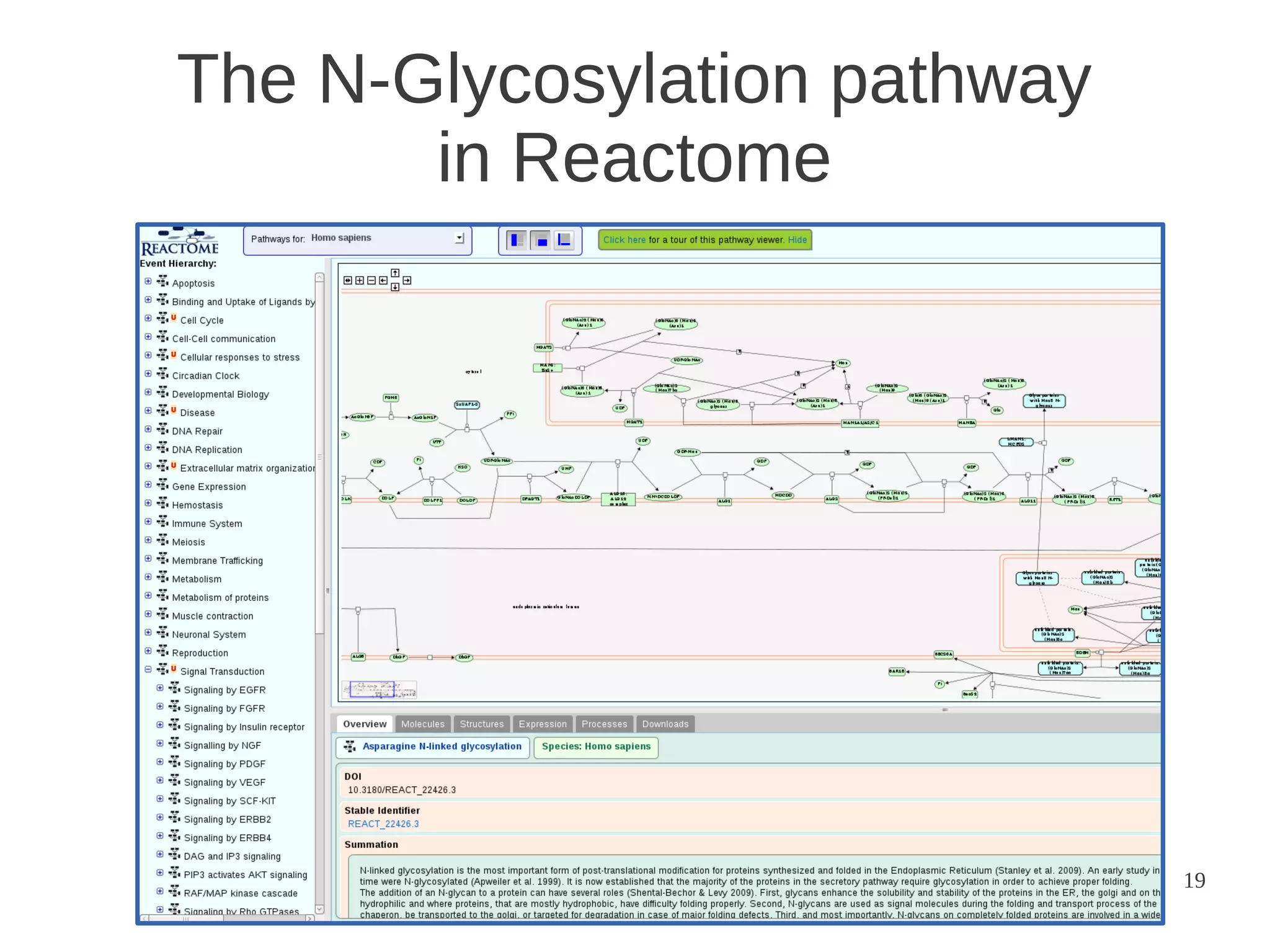
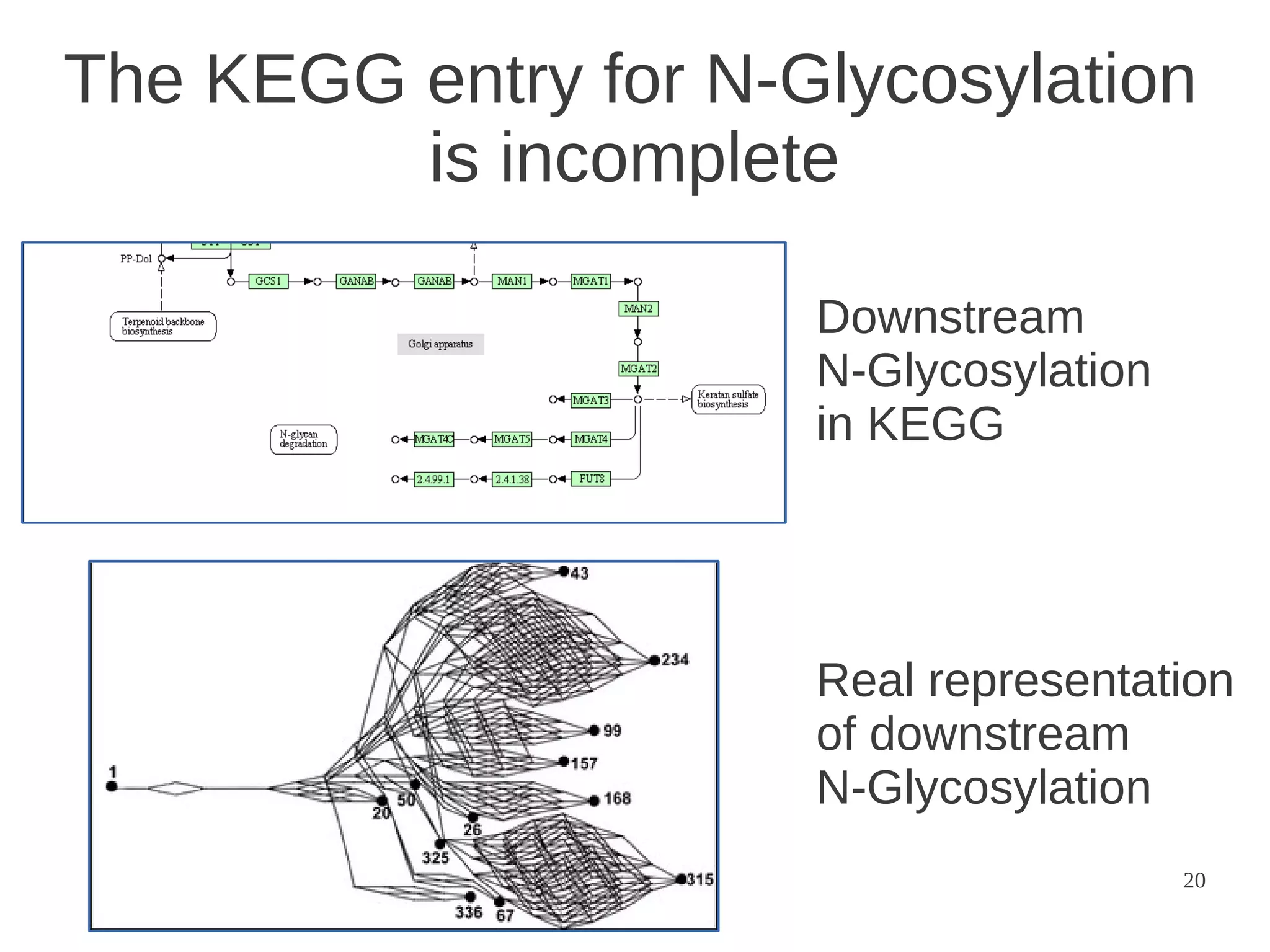
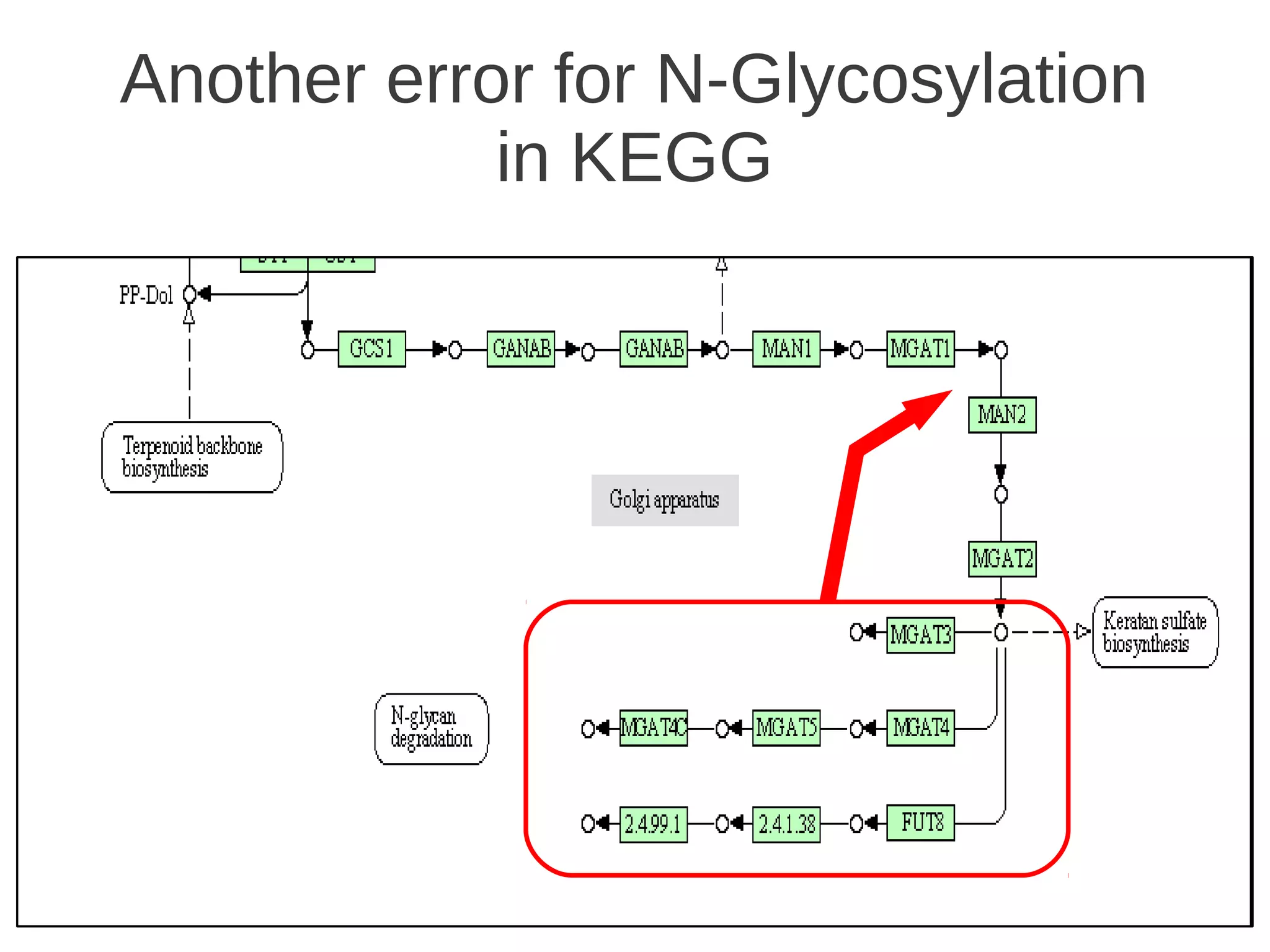
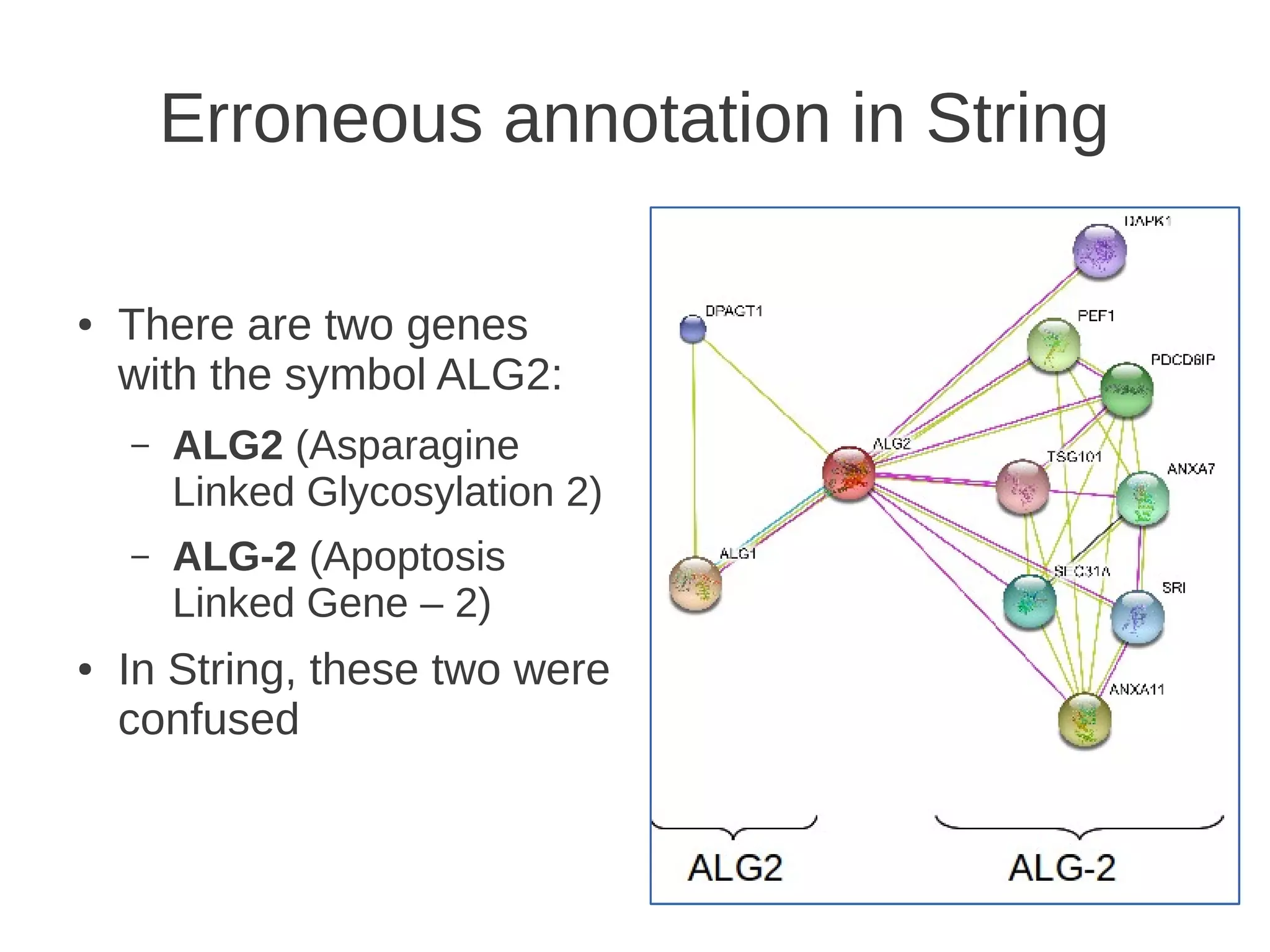
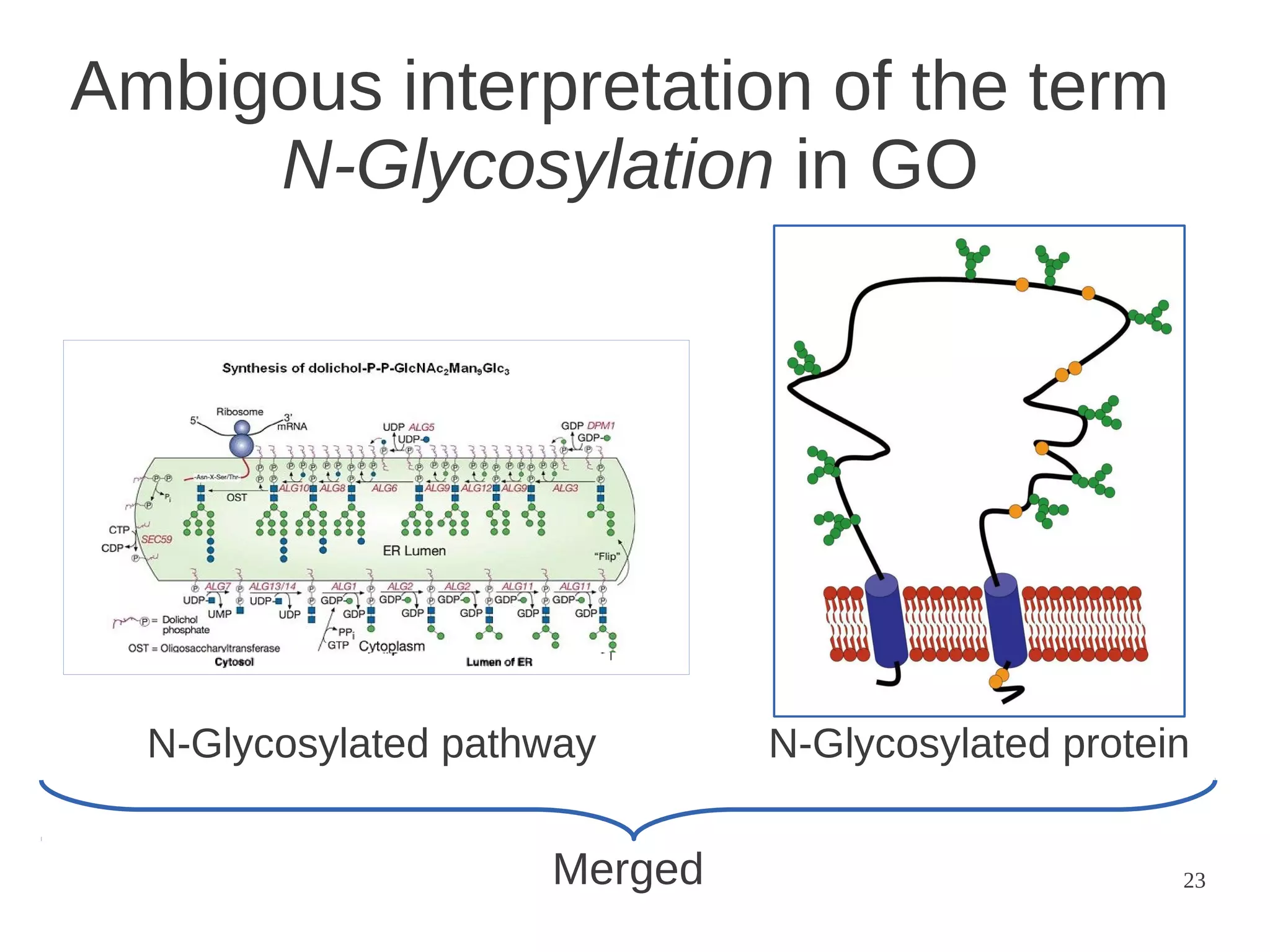
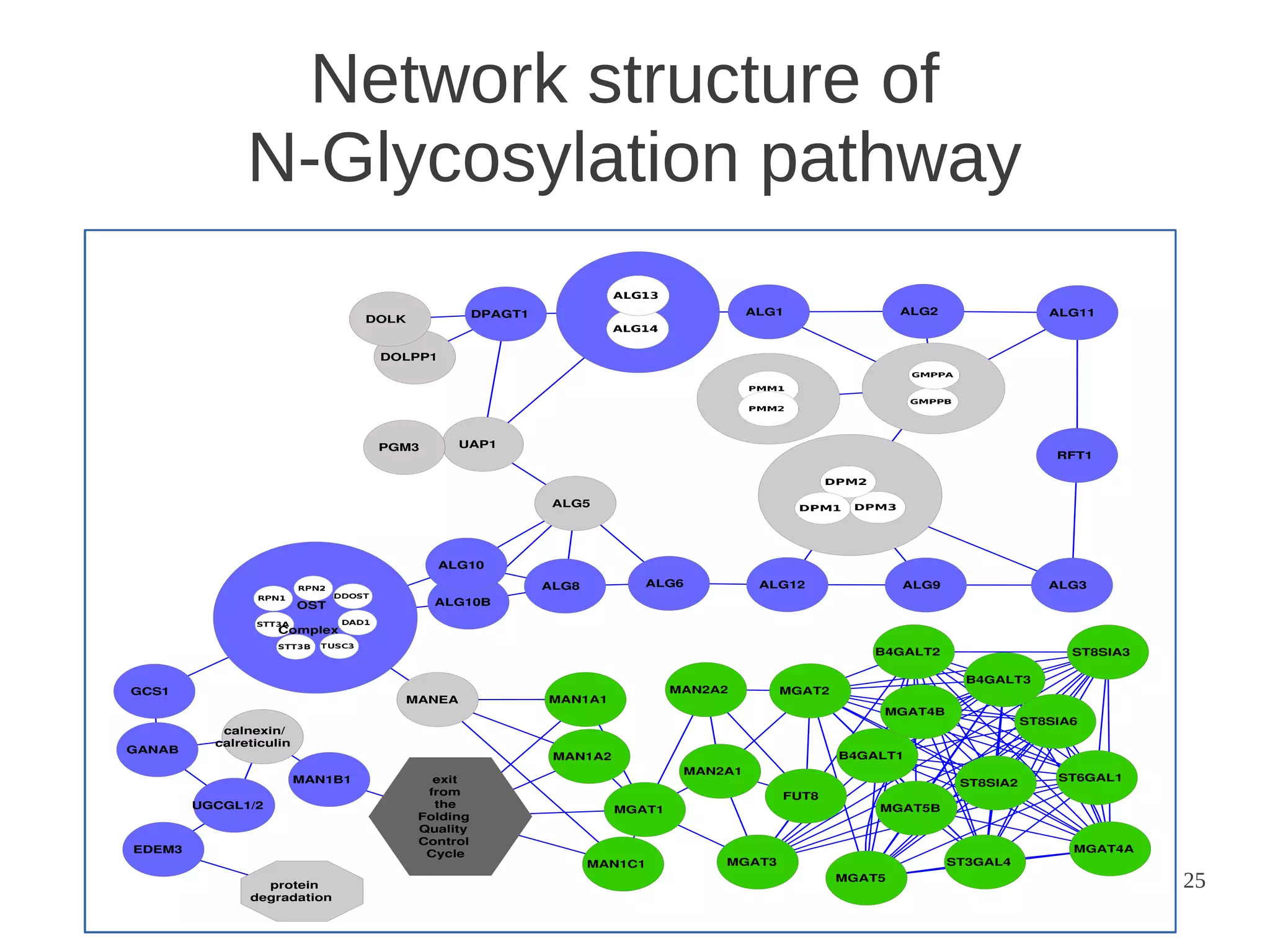
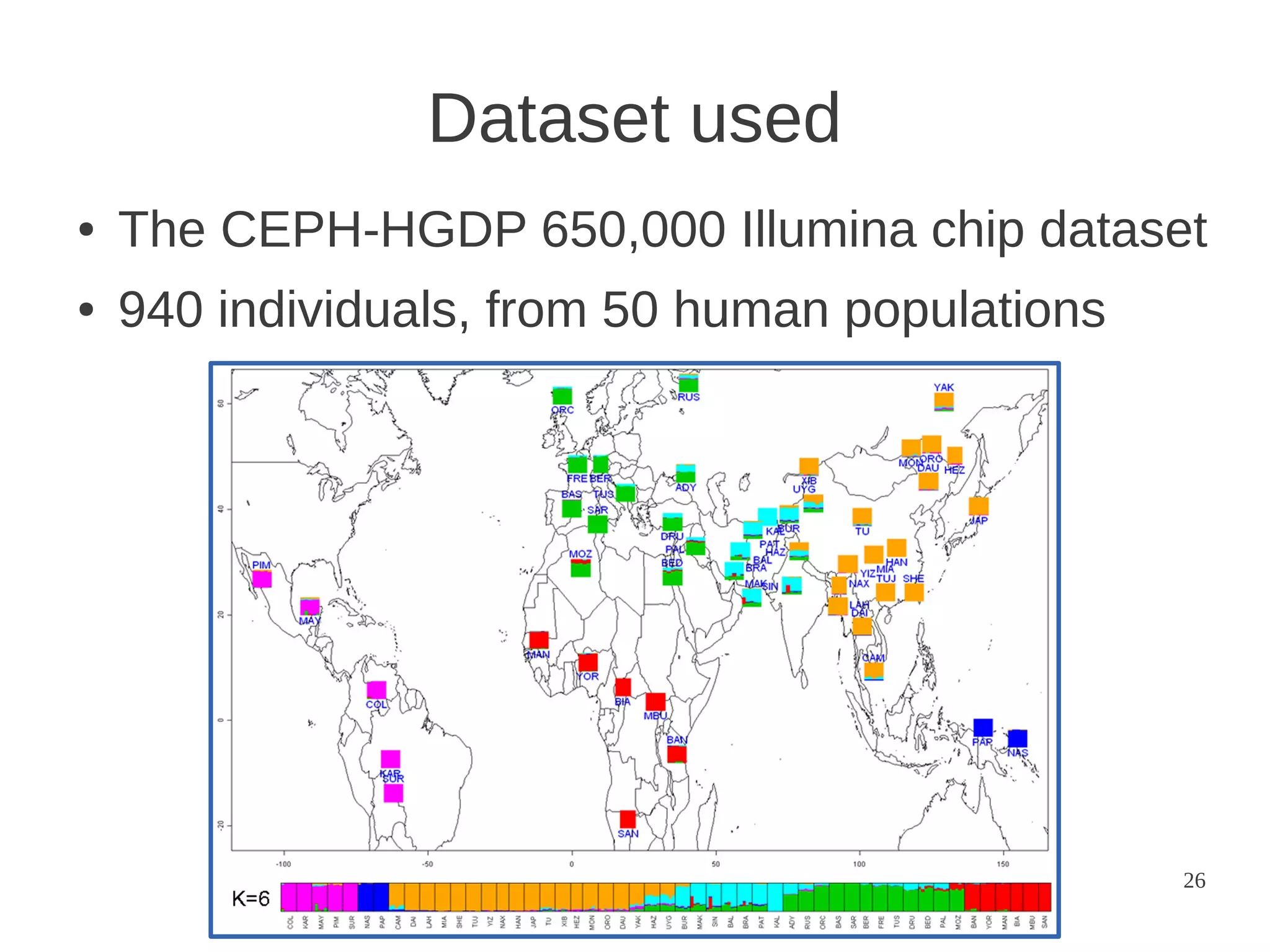
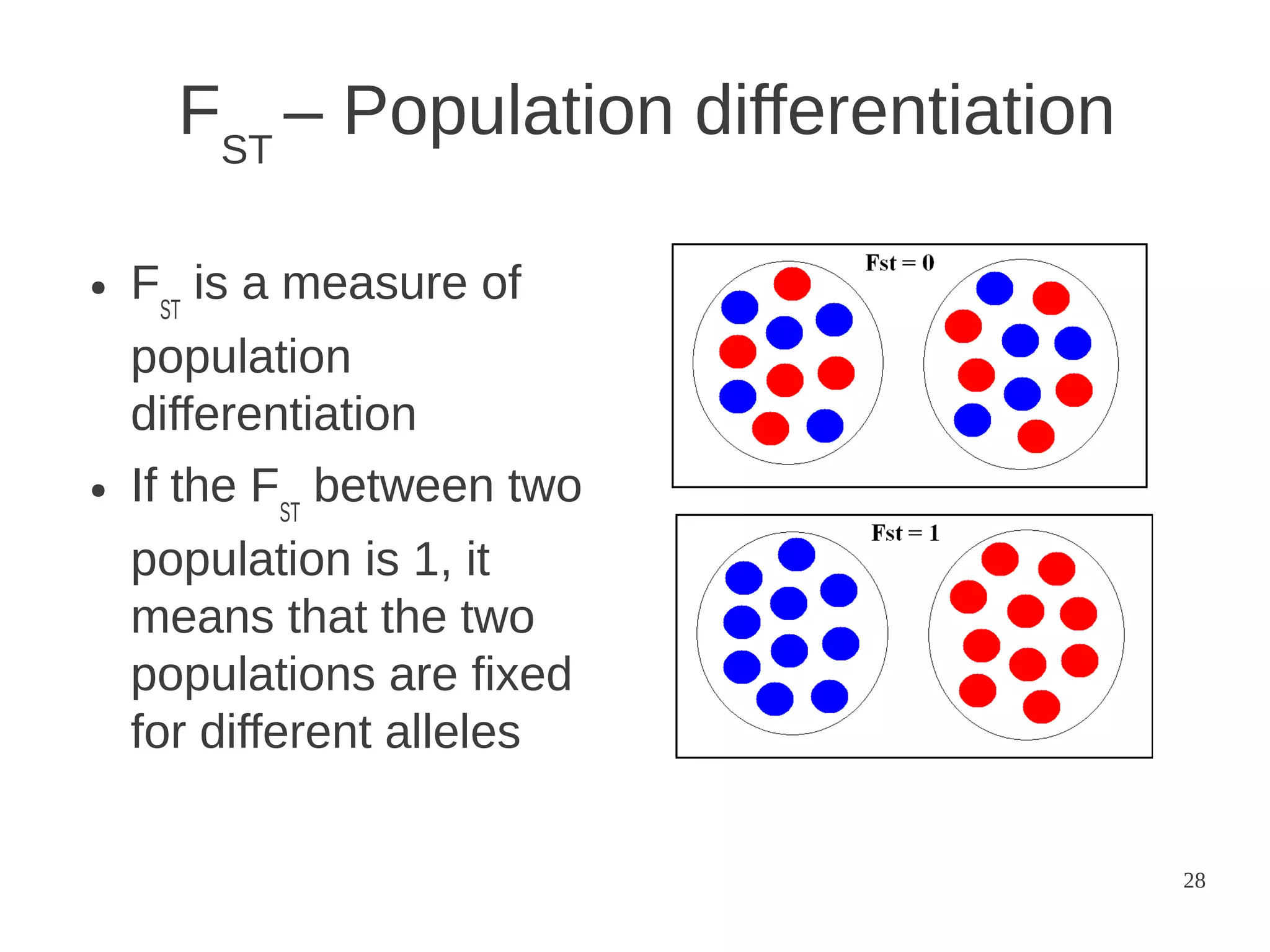
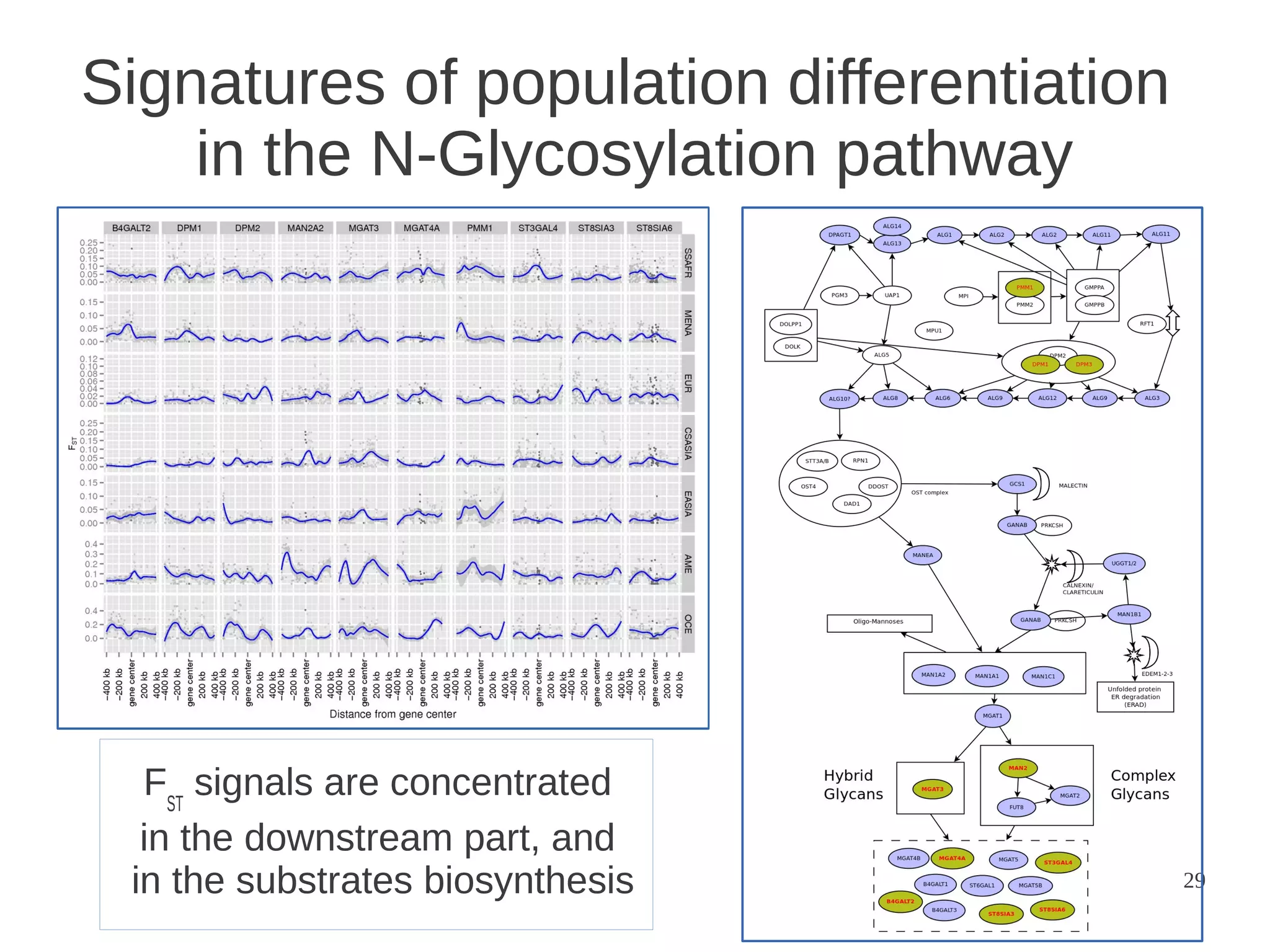
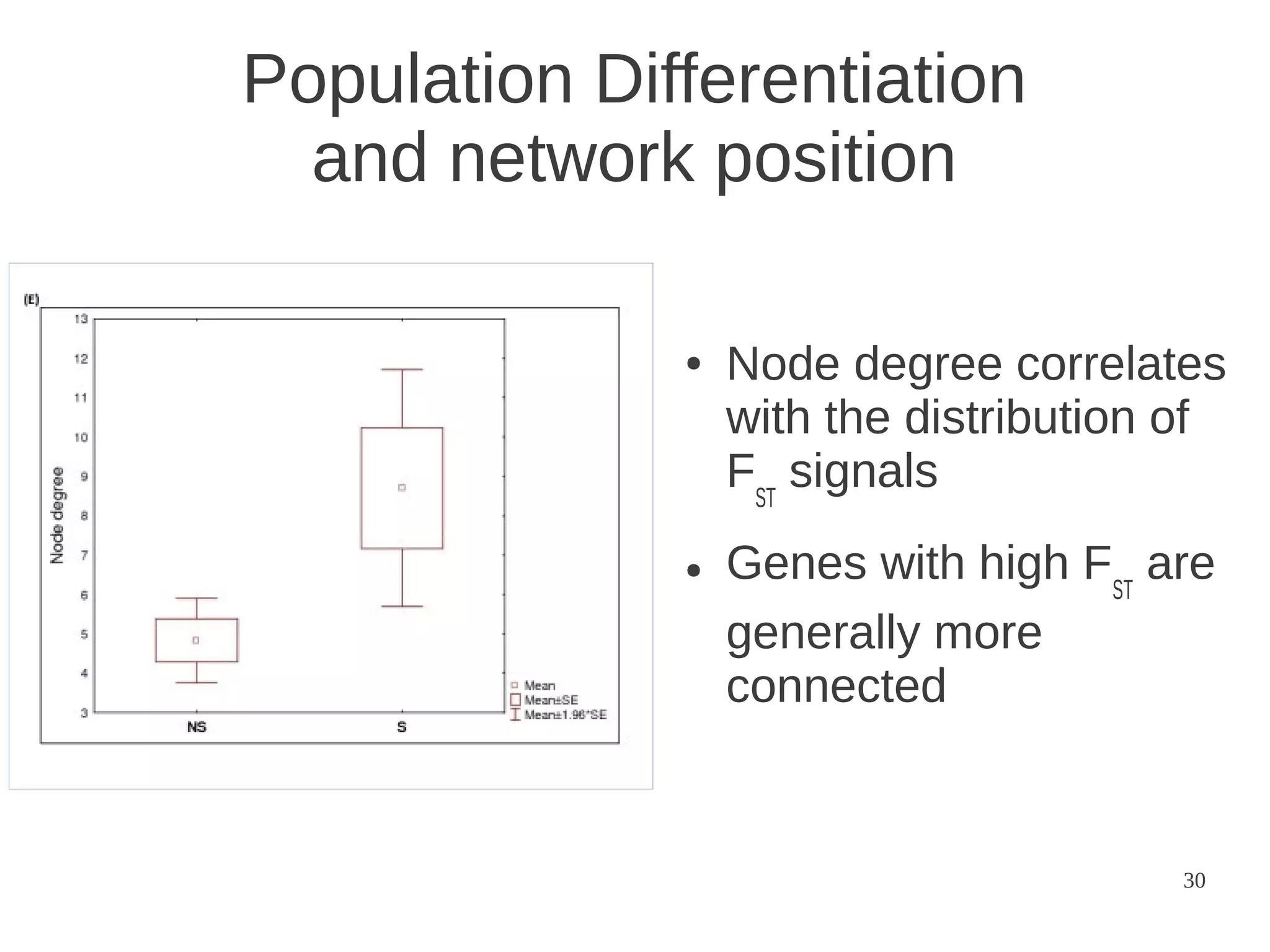
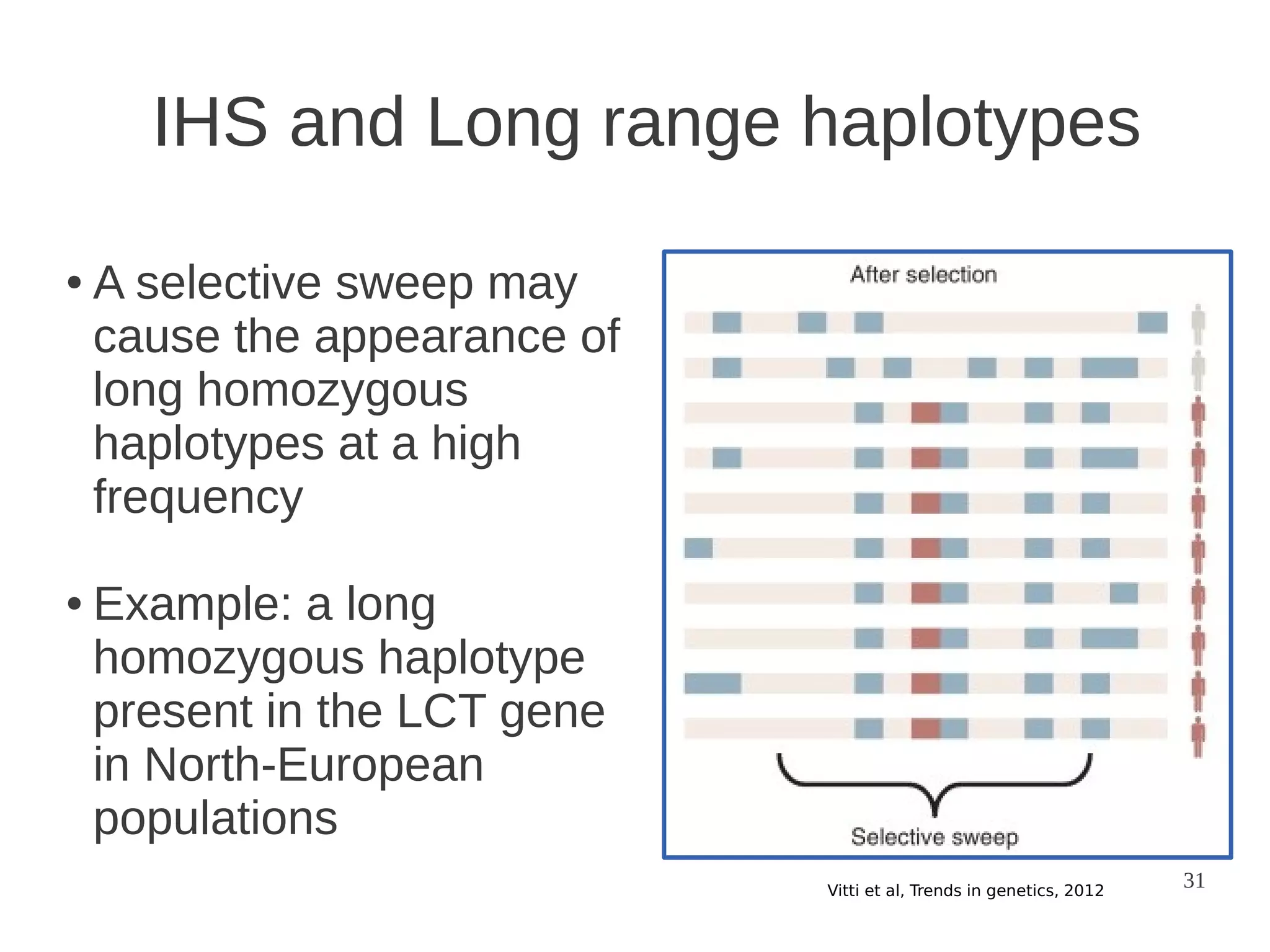
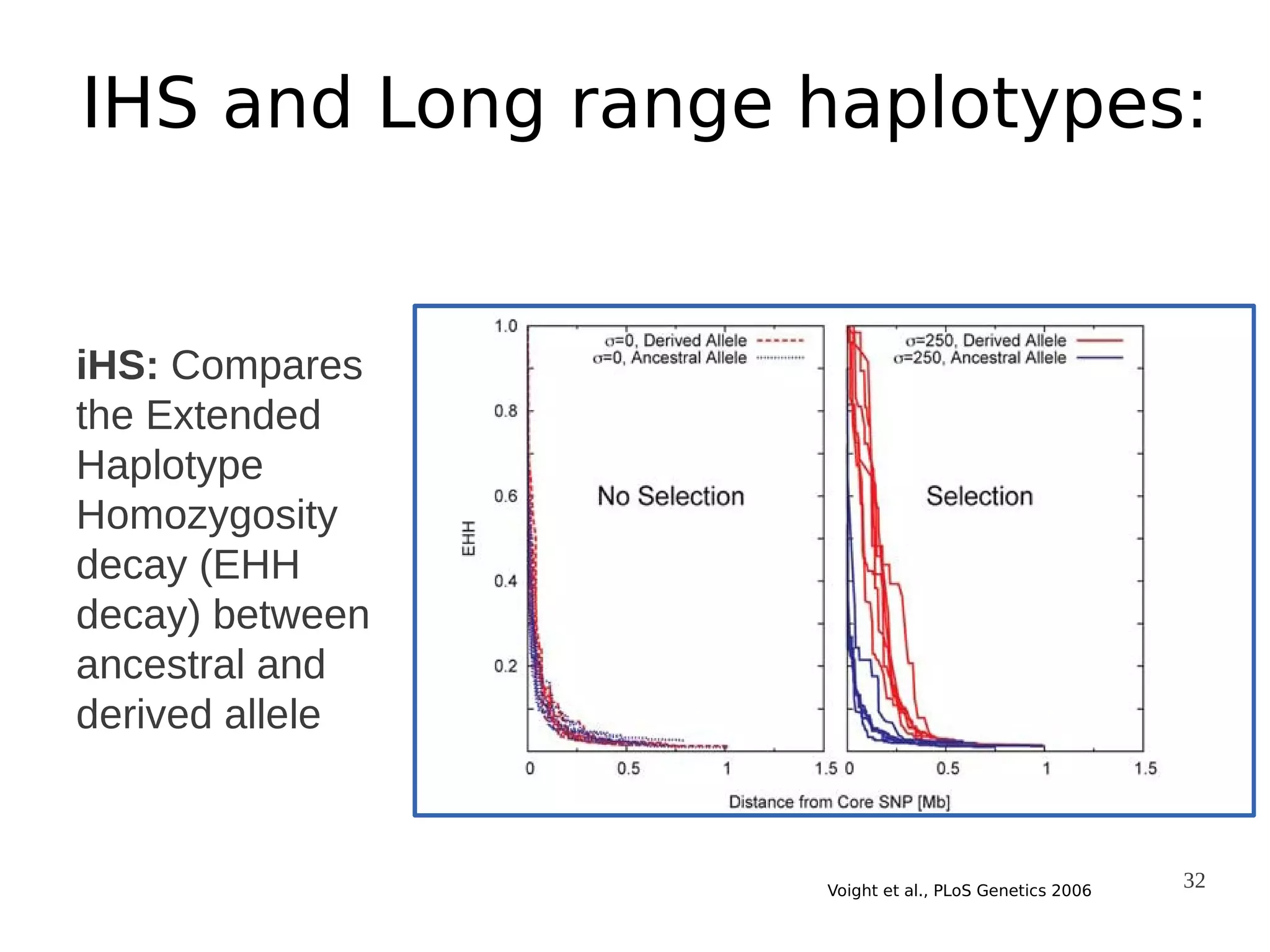
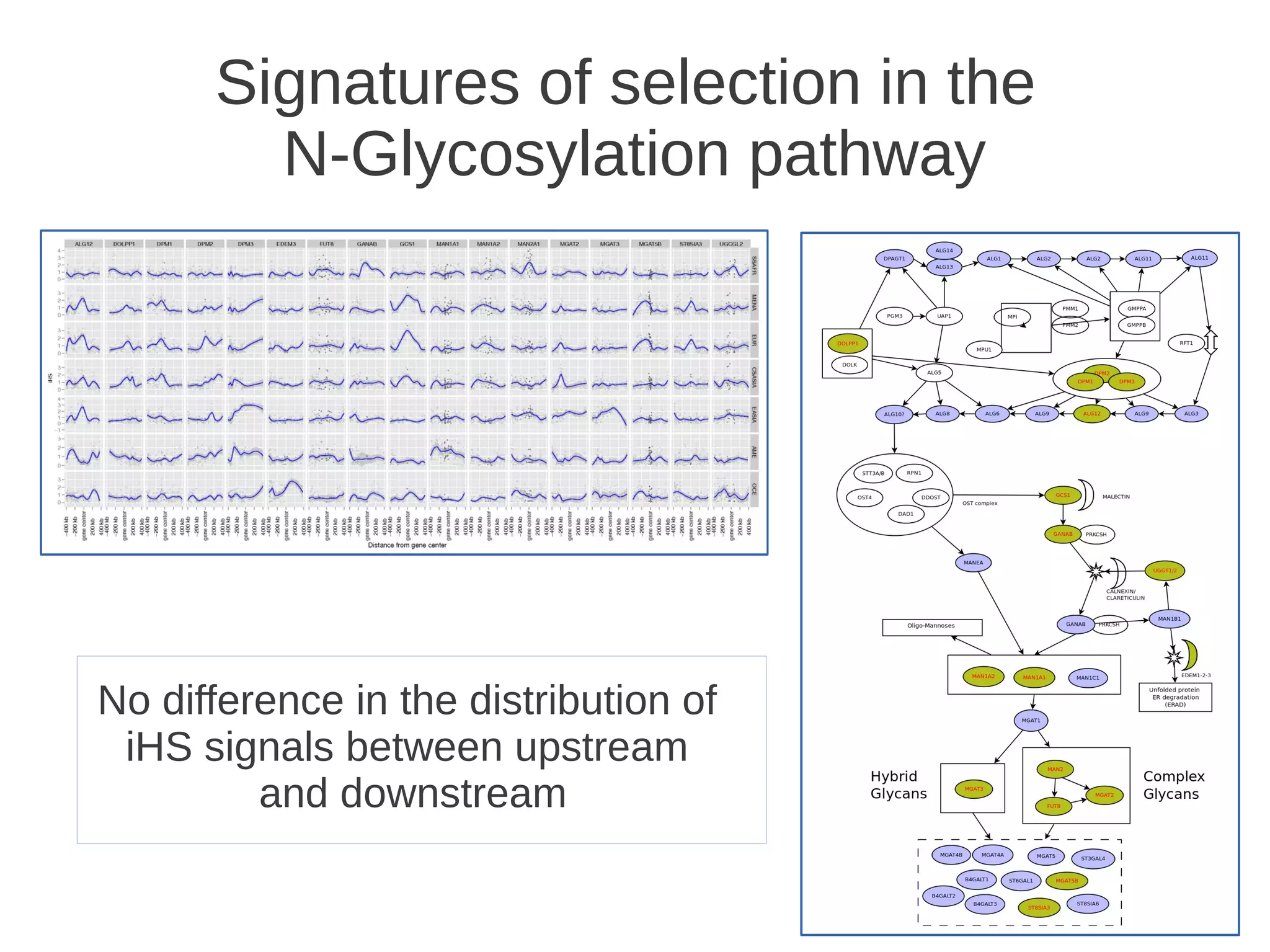
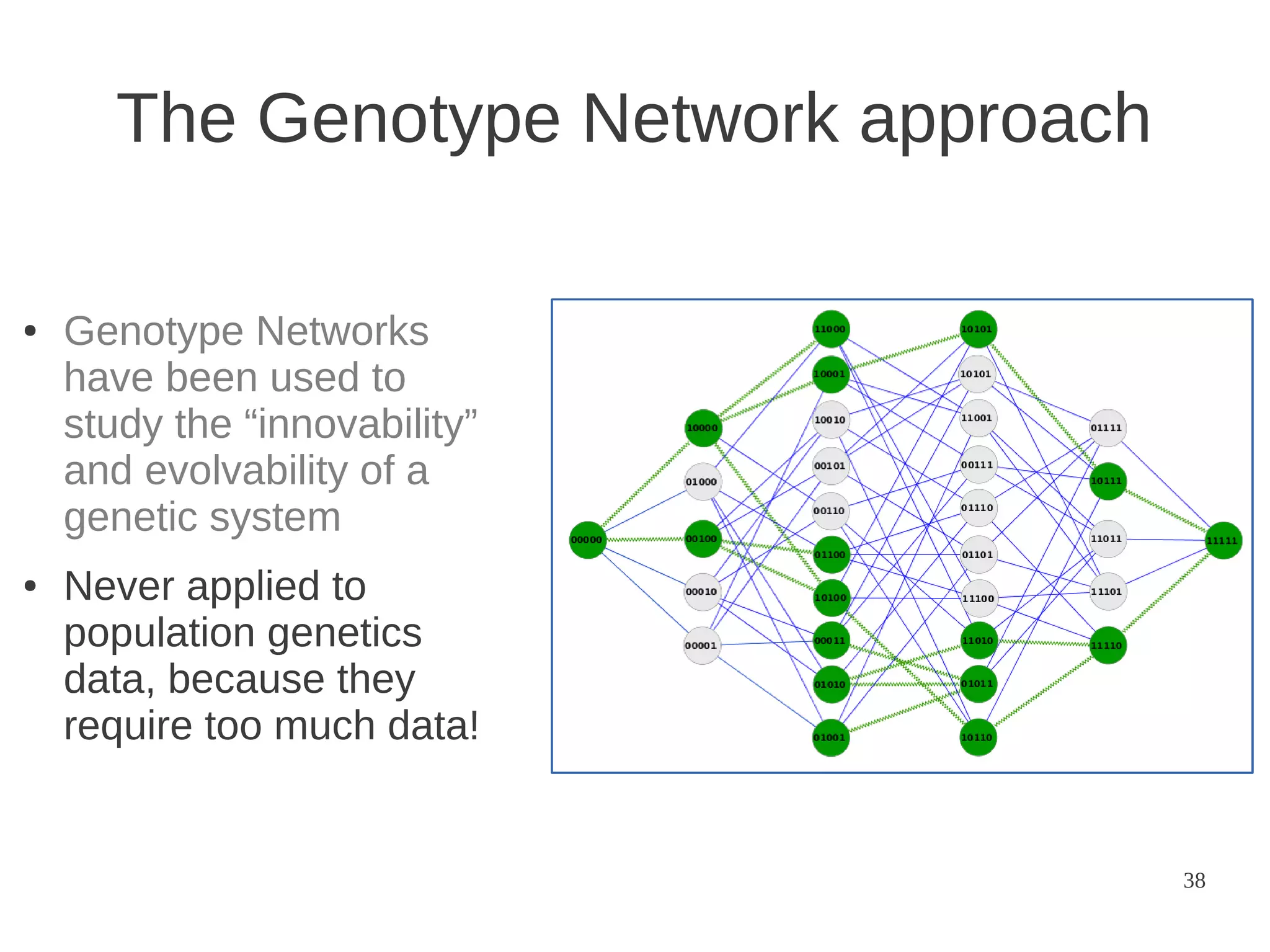
The document explores the application of network theory to human population genetics, focusing on the n-glycosylation pathway and genotype networks. It provides insights into genetic adaptation signals, population differentiation, and the development of resources for bioinformatics, including the Human Selection Browser and VCF2space. Key findings indicate that downstream parts of the n-glycosylation pathway show more genetic signatures of selection, while coding regions have higher connectivity in genotype networks compared to non-coding regions.











![Genotype Networks - theory
●
John Maynard-Smith:
the concept of a Protein
Space, which is explored
by populations
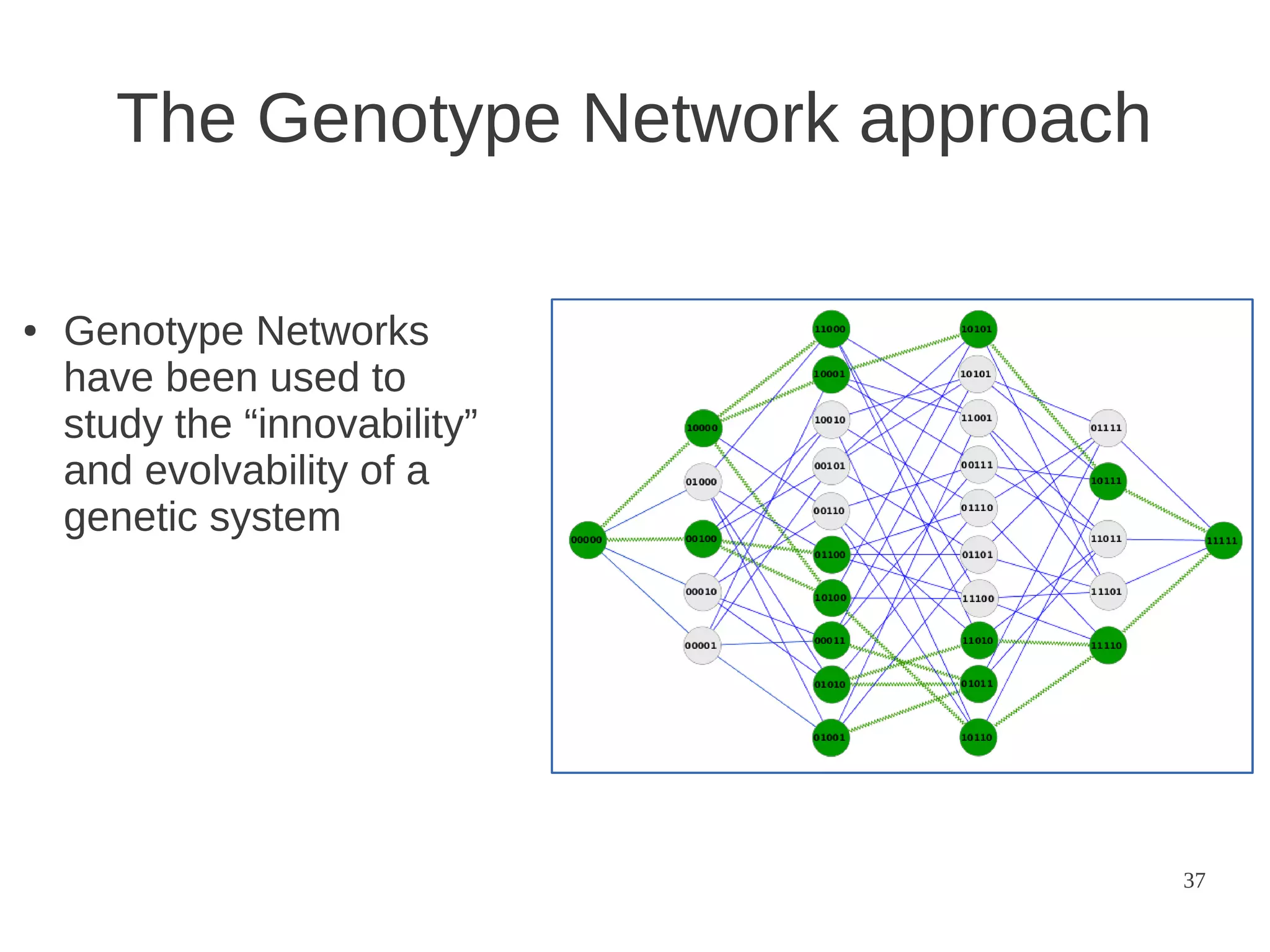
“if evolution by natural selection is
to occur, functional proteins [or
DNA sequences] must form a
continuous network which can be
traversed by unit mutational steps
without passing through nonfunctional intermediates”
40](https://image.slidesharecdn.com/dallolio-20gm-20-20thesis-20defense-131106044035-phpapp01/75/Thesis-defence-of-Dall-Olio-Giovanni-Marco-Applications-of-network-theory-to-human-population-genetics-from-pathways-to-genotype-networks-40-2048.jpg)